Vitamin D is a potent immunonutrient that through its main metabolite calcitriol, regulates the immunomodulation of macrophages, dendritic cells, T and B lymphocytes, which express the vitamin D receptor (VDR), and they produce and respond to calcitriol. Genetic association studies have shown that up to 65% of vitamin D serum variance may be explained due to genetic background.
- vitamin D
- polymorphism
- calcidiol
- calcitriol
- VDBP
- GC
- CYP2R1
- CYP27B1
- VDR
1. Introduction
The etiology and progression of autoimmune diseases (AIDs) are multifactorial and complex[1]. Genetic and environmental factors such as nutrients have been proposed to partially explain the pathophysiology progression of autoimmunity[2]. Notably, vitamin D regulates the growth and differentiation of various cells of the immune system such as macrophages, dendritic cells, T cells, and B cells, which are able to express the vitamin D receptor (VDR), produce and respond to the active form of vitamin D, calcitriol (1α,25(OH)
D
)[3].
In autoimmune diseases, epidemiological studies have reported a high prevalence of vitamin D deficiency by the quantification of calcidiol; this deficiency has been associated with worse disease clinical activity and progression of systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), and multiple sclerosis (MS) diseases[4]. Likewise, several clinical trials and murine studies using cholecalciferol supplementation have described vitamin D′s immunomodulatory properties in autoimmune diseases[5]. Lower calcidiol serum levels are a strong predictor for worse MS clinical manifestations [6] as well as RA patients show lower calcidiol levels than control subjects (CS), and they have shown a negative association of calcidiol serum levels with clinical disease activity[7]. Regarding SLE patients, a higher prevalence of calcidiol serum deficiency than the general population has also been reported[8], and the lower serum calcidiol in SLE patients is associated with high clinical disease activity[9].
Calcidiol serum deficiency in patients with autoimmune diseases and the general population could be attributed to several factors, including lack of exposure to sunlight, skin pigmentation, sunscreen use, nutrient intake deficiencies, age, use of glucocorticoids, and the genetic background of the populations[10]. The potential roles of 35 genes that could modulate the vitamin D serum levels status have been reported in previous studies, highlighting that multiple single nucleotide polymorphisms (SNPs) in these genes are associated with lower calcidiol serum levels[11], such as the SNPs described in the vitamin D binding protein (VDBP; rs2282679
), 25-hydroxylase (rs10751657,
), 1α-hydroxylase (rs10877012,
) and the vitamin D receptor (
(rs2228570),
(rs1544410),
(rs7975232), and
(rs731236)
). Therefore, the aim of this comprehensive literature review was to discuss the current findings of the functional SNPs in
,
,
, and
related to genetic risk and the most common clinical features of MS, RA, and SLE.
2. Vitamin D Status and Genetic Evidence in the Populations
Regarding vitamin D deficiency, several factors have been described, mainly the lack of exposure to sunlight, latitude, the season of the year, skin pigmentation, and use of sunscreen; other factors involved in vitamin D deficiency are diet, age, pharmacotherapy administered (antiepileptic and glucocorticoids), and particularly, several studies have described that genetic differences between individuals and populations such as genetic polymorphisms could influence the vitamin D deficiencies presented in all populations around the world[10].
Multiple SNPs in
,
,
, and
genes are associated with lower calcidiol serum levels[11]. Moreover, vitamin D serum deficiencies are present in a high frequency in a healthy population, which could be related to the SNPs’ presence in these genes that may modify the response to supplementation of vitamin D in health and disease. A study conducted in healthy Iranian adolescents described the differential effect of the
(rs10741657) A > G SNP on the supplementation of 50,000 UI of cholecalciferol weekly over 9 weeks, and showed that participants carrying the AA genotype presented 2.5-fold higher calcidiol serum levels in comparison to those that carrying the GG genotype (OR = 2.5 (1.4–4.4);
0.002)[12]. This is evidence of the role of polymorphisms in genes related to vitamin D metabolism in the variation of the response to vitamin D supplementation, even in healthy conditions.
3. Polymorphisms in the Main Key Genes Related to Vitamin D Metabolism
The skin produces around 80% of vitamin D in the form of cholecalciferol when exposed to ultraviolet B (UVB) light at a wavelength of 290–320 nm, and the remaining 20% is obtained from the diet as ergocalciferol from mushrooms or as cholecalciferol from fortified dairy products, fish, and eggs, mainly [13].
In blood, cholecalciferol and ergocalciferol bind to the vitamin D binding protein (VDBP), a protein encoded by the
gene to be transported to the liver (
1a). The
(rs2282679) SNP was been associated with modulation of vitamin D serum levels in several populations[14][15][16]. In these studies, the main vitamin D circulating metabolite in blood, calcidiol, was used to evaluate vitamin D deficiencies [17].
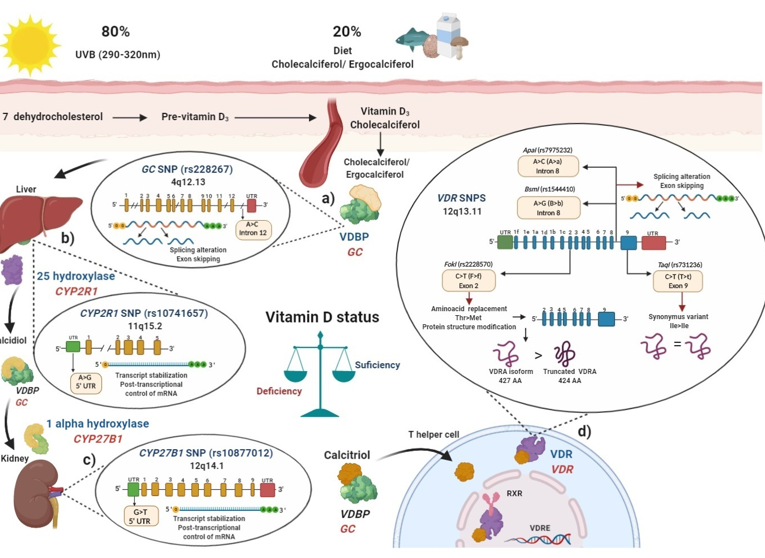
Polymorphisms in main key enzymes and proteins associated with vitamin D metabolism: localization and functional effects: (
) Vitamin D binding protein (VDBP) (encoded by
gene) binds to ergocalciferol/cholecalciferol in order to be transported to the liver;
(rs2282679) single nucleotide polymorphisms (SNP) due to its location in intron may generate a splicing alteration and exon skipping; (
) In the liver, 25 hydroxylase (encoded by
gene) converts ergocalciferol and cholecalciferol to calcidiol and then calcidiol binds to VDBP to be transported to the kidney;
(rs10741657) SNP located on the 5′ untranslated region (UTR) region may affect the transcript stabilization and the post-transcriptional control; (
) In the kidney, calcidiol is converted to calcitriol by the enzyme 1 alpha hydroxylase (encoded by the
gene);
(rs10877012) SNP located on 5′ UTR may affect the transcript stabilization and the post-transcriptional control of mRNA; (
) After calcitriol enters target cells and binds to vitamin D receptor (VDR) (encoded by
gene). Then, the VDR-calcitriol complex in the cytosol is translocated to the nucleus, where it binds to retinoid X receptor (RXR) to form a heterodimer, which interacts with vitamin D response element (VDRE) in vitamin D target genes, i.e., in T helper (Th) lymphocytes to suppress
or activate
Mainly four SNPs have been described in the
gene: the
(rs2228570) located on exon 2, which generates a non-synonymous polymorphism with a change of C > T (also called F > f) and this results in a change of threonine to methionine. The presence of the restriction site
C allele (F allele), generates a new start codon (ATG) 9 bp after of the common starting site, which translate to an shorter truncated VDR protein of 424 amino acids with more transactivation capacity as a transcription factor than the wild type full-length VDR A isoform (VDRA) of 427 amino acids; the
(rs1544410) located on intron 8 presents a change of A > G (also called B > b), could affect messenger ribonucleic acid (mRNA) stability and the gene expression of
, and also it could generate an alteration in the splice sites for mRNA transcription or a change in the intron regulatory elements of
;
(rs7975232) located on intron 8 of
presents a change of A > C (also called A > a), does not change the amino acid sequence of the VDR protein, therefore could affect mRNA stability and the gene expression of
;
(rs731236) is located on the exon 9 of
, presents a change of C > T (also called T > t) and generates a synonymous change of the isoleucine amino acid in the coding sequence, therefore it does not change the encoded protein, but it could influence the stability of the mRNA. All these SNPs are related to modulating de vitamin D serum status in health and disease. Ile: isoleucine; Thr: threonine; Met: methionine VDBP: vitamin D binding protein; VDR: vitamin D receptor; RXR: retinoid X receptor; VDRE: vitamin D response elements; UTR: untranslated region; THEM4: thioesterase superfamily member 4; Th: T helper lymphocyte; VDRA: wild type full-length VDR A isoform.
In the liver, cholecalciferol and ergocalciferol are converted to calcidiol by the enzyme vitamin D 25-hydroxylase which is encoded by the
gene (
1b), the (rs10741657) SNP described in
had also been related to modulate calcidiol serum status in several studies[12][14][15][18]. Then, after the generation of calcidiol in the liver, it binds again to VDBP and subsequently interacts with the enzyme 1-α hydroxylase, encoded by the
gene, mainly in the proximal kidney tubule, calcidiol is converted to calcitriol (1α,25 dihydroxyvitamin D), which is the biological and functional active form of vitamin D (
, particularly the rs10877012 SNP, was associated with lower calcidiol serum levels[19].
Once the generation of calcitriol is performed, it is bound to VDBP and is transported to target cells and tissues where in cytoplasm or membrane, it interacts with the vitamin D receptor (VDR), encoded by the
gene.
After the calcitriol translocation to the nucleus, this complex forms a heterodimer with the retinoid x receptor (RXR), and thus regulates vitamin D target genes through its binding to specific DNA sequences called vitamin D response elements (VDREs)[20][21]. Since VDR is expressed in different immune cells, such as neutrophils, macrophages, dendritic cells (CDs), T and B lymphocytes, calcitriol may regulate the immune system[8].
Calcitriol can be produced by monocytes and macrophages and generate a shift from pro-inflammatory to tolerogenic immune status[22]. Calcitriol promotes M1 phenotype switching to M2 via the nVDR-PPARγ pathway and via the upregulation of the expression of IL-10[23]. Particularly, calcitriol promotes a shift from Th1 and Th17 to Th2 immune profile via suppression of expression of cytokines of the Th1 (IL-2, IFN-γ, TNF-α) and Th17 (IL-17, IL-21) profiles, and induction of the expression of cytokines of Th2 profile (IL-4, IL-5, IL-9, IL-13), in order to limit inflammatory processes and autoimmune reactions [8][22].
Several studies have focused on evaluating SNPs described in the
, such as
(rs2228570),
(rs1544410),
(rs7975232), and
(rs731236) polymorphisms, which was associated in several populations with lower calcidiol levels (
1d). Therefore, SNPs′ presence in key enzymes related to vitamin D metabolism may modulate calcidiol levels and calcitriol function; they may also modify the disease activity in MS, RA, and SLE through the vitamin D deficiency, and contribute to genetic susceptibility to autoimmunity. In the following sections, we will describe how polymorphisms in these genes can modulate their expressions.
3.1. Vitamin D Binding Protein (VDBP) (SNP rs2282679
3.1. Vitamin D Binding Protein (VDBP) (SNP rs2282679
GC)
)
Vitamin D binding protein (VDBP) is the main carrier protein for vitamin D, which binds 85 to 90% of the total circulating calcidiol [24]. This protein is encoded by the
gene located in chromosome 4, position 13.3 (4q13.3), and has 55,136 bp size [25]. The
gene consists of 13 exons and the rs2282679 SNP was described located at the position 71,742, presenting an A ˃ C change (ancestral allele: A) in the intron 12, near to the actin III subdomain[26] and the endonuclease enzyme commonly used to identify this
(rs2282679) SNP is
(
) [27]. According to rs2282679 SNP intron location, its hypothetical functional effect may alter genetic mRNA expression due to modification of the splicing process (44). A complete GWAS showed that
(rs2282679) C allele was associated with lower serum calcidiol and VDBP levels in a study carried out in approximately 30,000 subjects with European ancestry included in 15 cohorts[28]. Additionally, various studies have shown the association of the
(rs2282679) SNP with genetic susceptibility and disease modulation in MS, RA, and SLE[29][30][31][32] (
1a).
3.2. Vitamin D 25-Hydroxylase (SNP rs10741657
3.2. Vitamin D 25-Hydroxylase (SNP rs10741657
CYP2R1)
)
The enzyme vitamin D 25-hydroxylase is a protein of around 500 amino acids with a molecular weight of 50–55 kilo-daltons (kDa). The liver is the main site where it is synthesized and performs its function of 25-hydroxylation enzyme activity, besides this enzyme activity was also described in the kidney and intestines[33], but cholecalciferol and ergocalciferol are metabolized mainly by the vitamin D 25-hydroxylase to calcidiol in the liver[34].
The enzyme vitamin D 25-hydroxylase is encoded by the
gene located in chromosome 11 in the short arm at position 15.2 (11p15.2) and consists of 15,500 bp. The SNP rs10741657
is positioned at 14,893,332 pb in the gene, it displays an A ˃ G change in the 5′ UTR region (ancestral allele: A)[18].
In order to recognize the presence of this SNP, the endonuclease restriction enzyme
(
) is commonly used[35]. According to the SNP location in 5′ UTR, the rs10741657 SNP may regulate gene expression by modifying translation initialization and transcript stabilization of mRNA, and therefore, modulate 25-hydroxylase expression and enzymatic activity rate[36].
3.3. Vitamin D 1-α Hydroxylase (SNP rs10877012
3.3. Vitamin D 1-α Hydroxylase (SNP rs10877012
CYP27B1)
)
The enzyme vitamin D 1-α hydroxylase corresponds to a P450 protein of 507 amino acids of around 55 kDa[37]. This enzyme metabolizes calcidiol to calcitriol, the active form of vitamin D[34], and it is encoded by the
gene, which is found in chromosome 12, long arm, position 14.1 (12q14.1), its size is 6653 bp.
The SNP rs10877012
is located at the non-coding region -1260 and is characterized by a change of G ˃ T (ancestral allele: G)[38][39]. The restriction endonuclease enzyme
(
is commonly used for its detection [27][35]. Due to its location in the 3′ UTR region, this SNP may alter transcript stabilization regulation and its localization in the cytoplasm [36].
A study carried out in 253 German patients with differentiated thyroid carcinoma showed that patients with the presence of the GG genotype (Referred as CC in this study by its position in the negative DNA strand) was associated with lower calcitriol serum levels than patients carrying the TT genotype (Referred as AA in this study by its position in negative DNA strand) (60 pmol/mL vs. 72 pmol/mL, respectively)[38].
In another study carried out in Caucasian German patients with gestational diabetes, the GG (CC) genotype was also associated with lower calcidiol levels[19]. Likewise, another study carried out in a healthy Caucasian British population demonstrated an association between the
(rs10877012) G (C) allele and lower serum calcidiol levels[40].
Besides, in type 1 diabetes in Caucasian patients from Germany, those carrying the GG (CC) genotype had a reduced amount of mRNA from
compared to HS (1.6855 vs. 1.8107, respectively,
= 0.0220)[19]. Therefore, the SNP rs10877012
may also modulate the vitamin D serum status and the genetic susceptibility or disease modulation in autoimmune diseases such as MS, RA, and SLE (
1c).
3.4. Polymorphisms in Vitamin D Receptor (VDR)
Most genetic studies evaluating the potential association of calcidiol serum levels with genetic polymorphisms have focused on evaluating the polymorphisms described in the vitamin D receptor (VDR). The VDR is a member of the steroid/thyroid hormone receptor superfamily; and this receptor is encoded by the gene with the same name,
, which is located in chromosome 12, position 12q.13.11, comprising a region of approximately 100,000 bp of DNA, and only 4600 bp encode the VDR protein.
Functionally VDR is a transcription factor regulated by ligand binding and possibly by phosphorylation events[41]. It is a soluble 427 amino acid protein located mainly in the nucleus, cell cytoplasm, and cellular membrane, from where it translocates to the nucleus through the microtubule system after interaction with its ligand, calcitriol [42].
VDR is expressed in various organs involved in calcium metabolism, immune cells, and the nervous system (5). Three isoforms of the VDR were described.
The most common is the VDRA isoform of 427 amino acids and 48 kDa, with a start site in exon 2. The second is a long VDRB1 isoform of 477 amino acids and 54 kDa, this isoform presents 50 amino acids more in the N-terminal domain by an ATG start site in the exon 1d, described in the human kidney as well as in intestinal and renal epithelial cell lines[42][43].
The third is a shorter VDRA isoform of 424 amino acids and greater transactivation capacity as a transcription factor, caused by the SNP
in the exon 2[42].
More than 14 different polymorphisms were described in human
, which could influence the modulation of the response to calcitriol by binding to VDR. The four SNPs most frequently studied are:
(rs2228570),
(rs1544410),
(rs7975232), and
(rs731236) [41][44]. These were related to modulating the vitamin D status independently (in alleles and risk genotypes) as well as in haplotypes and haplogenotypes, both in original articles and meta-analyses (
1d).
3.4.1. FokI (rs2228570) VDR SNP
(rs2228570)
SNP, also referred as the start codon polymorphism (SCP), was defined using the
) restriction enzyme in a restriction fragment length polymorphism test (RFLP) [45].
(rs2228570) is located in exon 2 and is considered a non-synonymous polymorphism, because the change of C > T (ancestral allele T), also referred as F > f change, which generates a non-synonymous change of threonine to methionine and dictates two potential translation initiation sites[44].
The presence of the restriction site
is when the C allele is presented (also called F allele by the cut of the
restriction enzyme), this C allele generates a new start codon (ATG) 9 bp after the common starting site, which translates to a shorter VDRA protein of 424 amino acids instead of the wild type full-length VDRA isoform of 427 amino acids[45].
SNP (rs2228570) was found to be functional, and the short 424 amino acid VDRA isoform is somewhat more active than the long VDRA isoform of 427 amino acid, in terms of its transactivation capacity as a transcription factor[46][47]. In the absence of the restriction site
, the T allele (also called f allele), translation begins at the first original site at the exon 2, and the VDRA of 427 amino acids is expressed, which is 1.7-fold less active in its transactivation capacity, and presents less stability[48][49][50][51].
3.4.2. BsmI (rs1544410) and ApaI (rs7975232) VDR SNPs
(rs1544410) and
(rs7975232)
SNPs were defined using the
(
) and the
(
restriction enzyme, respectively in a RFLP test.
(rs1544410)
SNP, located in the intron 8, presents a change of A > G (also called B > b), and the ancestral allele is the G allele[49]. Regarding its functional effect, it could generate an alteration in the splice sites for mRNA transcription or a change in the intron regulatory elements of
.
SNP, also is located in the intron 8, presents a change of A > C, (also called A > a), and the ancestral allele is the C allele [21]. Both SNPs are located at the 3′ end of the
and do not change the amino acid sequence of the VDR protein. Therefore, they could affect mRNA stability and the gene expression of
3.4.3. TaqI (rs731236) VDR SNP
3.4.3. TaqI (rs731236) VDR SNP
(rs731236)
SNP, located in the exon 9, was defined using the
(
) restriction enzyme, and presents a change of C > T, (also called T > t), and the ancestral allele is the C allele.
generates a synonym change of the coding sequence; therefore, it does not produce an amino acid change of the encoded protein, but it could influence the stability of the mRNA [49].
If
(rs731236)
SNP is in high LD with
(rs7975232)
SNP, its functional effect is the possible modification of one of the zinc fingers of the nuclear signaling heterodimer that binds to the VDREs located in the target genes[52].
Due to the closeness of these four
SNPs, they were studied to determine their LD. In various populations of SLE, RA, and MS patients, the
(rs1544410),
(rs7975232), and
(rs731236)
SNPs were described with a strong LD[53][54], which infers that the alleles of these three polymorphisms could segregate into haplotypes from one generation of persons to another. In the case of
(rs2228570), it was described in a low LD with the other three
SNPs, which suggests that
does not segregate in blocks with others downstream
SNPs [53][54]. However, because the genetic recombination points vary between populations,
was also studied in haplotypes together with the
(rs1544410),
(rs7975232), and
(rs731236)
References
- Selmi, C.; Lu, Q.; Humble, M.C. Heritability versus the role of the environment in autoimmunity. J. Autoimmun. 2012, 39, 249–252.
- Floreani, A.; Leung, P.; Gershwin, M. Enviromental basis of autoimmunity. Clin. Rev. Allergy Immunol. 2016, 50, 287–300.
- Adorini, L.; Penna, G. Control of autoimmune diseases by the vitamin D endocrine system. Nat. Clin. Pract. Rheumatol. 2008, 4, 404–412.
- Arnson, Y.; Amital, H.; Shoenfeld, Y. Vitamin D and autoimmunity: New aetiological and therapeutic considerations. Ann. Rheum. Dis. 2007, 66, 1137–1142.
- Dankers, W.; Colin, E.M.; van Hamburg, J.P.; Lubberts, E. Vitamin D in Autoimmunity: Molecular Mechanisms and Therapeutic Potential. Front. Immunol. 2017, 20, 697.
- Duan, S.; Lv, Z.; Fan, X.; Wang, L.; Han, F.; Wang, H.; Bi, S. Vitamin D status and the risk of multiple sclerosis: A systematic review and meta-analysis. Neurosci. Lett. 2014, 570, 108–113.
- Lin, J.; Liu, J.; Davies, M.L.; Chen, W. Serum Vitamin D Level and Rheumatoid Arthritis Disease Activity: Review and Meta-Analysis. PLoS ONE 2016, 11, e0146351.
- Shoenfeld, Y.; Giacomelli, R.; Azrielant, S.; Berardicurti, O.; Reynolds, J.A.; Bruce, I.N. Vitamin D and systemic lupus erythematosus—The hype and the hope. Autoimmun. Rev. 2018, 17, 19–23.
- Sakthiswary, R.; Raymond, A.A. The Clinical Significance of Vitamin D in Systemic Lupus Erythematosus: A Systematic Review. PLoS ONE 2013, 8, e55275.
- Brouwer-Brolsma, E.M.; Vaes, A.M.M.; van der Zwaluw, N.L.; van Wijngaarden, J.P.; Swart, K.M.A.; Ham, A.C.; van Dijk, S.C.; Enneman, A.W.; Sohl, E.; van Schoor, N.M.; et al. Relative importance of summer sun exposure, vitamin D intake, and genes to vitamin D status in Dutch older adults: The B-PROOF study. J. Steroid Biochem. Mol. Biol. 2016, 164, 168–176.
- Sepulveda-Villegas, M.; Elizondo-Montemayor, L.; Trevino, V. Identification and analysis of 35 genes associated with vitamin D deficiency: A systematic review to identify genetic variants. J. Steroid Biochem. Mol. Biol. 2020, 196, 105516.
- Khayyatzadeh, S.S.; Mehramiz, M.; Esmaeily, H.; Mirmousavi, S.J.; Khajavi, L.; Salehkhani, F.N.; Hanachi, P.; Bahrami-Taghanaki, H.; Eslami, S.; Vatanparast, H.; et al. A variant in CYP2R1 predicts circulating vitamin D levels after supplementation with high-dose of vitamin D in healthy adolescent girls. J. Cell. Physiol. 2019, 234, 13977–13983.
- Christakos, S.; Dhawan, P.; Verstuyf, A.; Verlinden, L.; Carmeliet, G. Vitamin D: Metabolism, Molecular Mechanism of Action, and Pleiotropic Effects. Physiol. Rev. 2016, 96, 365–408.
- Slater, N.A.; Rager, M.L.; Havrda, D.E.; Harralson, A.F. Genetic Variation in CYP2R1 and GC Genes Associated with Vitamin D Deficiency Status. J. Pharm. Pract. Res. 2017, 30, 31–36.
- Nissen, J.; Vogel, U.; Ravn-Haren, G.; Andersen, E.W.; Madsen, K.H.; Nexø, B.A.; Andersen, R.; Mejborn, H.; Bjerrum, P.J.; Rasmussen, L.B.; et al. Common variants in CYP2R1 and GC genes are both determinants of serum 25-hydroxyvitamin D concentrations after UVB irradiation and after consumption of vitamin D3–fortified bread and milk during winter in Denmark. Am. J. Clin. Nutr. 2015, 101, 218–227.
- Cheung, C.-L.; Lau, K.-S.; Sham, P.-C.; Tan, K.C.; Kung, A.W. Genetic variant in vitamin D binding protein is associated with serum 25-hydroxyvitamin D and vitamin D insufficiency in southern Chinese. J. Hum. Genet. 2013, 58, 749–751.
- Holick, M.F. Vitamin D Status: Measurement, Interpretation, and Clinical Application. Ann. Epidemiol. 2009, 19, 73–78.
- Duan, L.; Xue, Z.; Ji, H.; Zhang, D.; Wang, Y. Effects of CYP2R1 gene variants on vitamin D levels and status: A systematic review and meta-analysis. Gene 2018, 678, 361–369.
- Ramos‐Lopez, E.; Kahles, H.; Weber, S.; Kukic, A.; Penna‐Martinez, M.; Badenhoop, K.; Louwen, F. Gestational diabetes mellitus and vitamin D deficiency: Genetic contribution of CYP27B1 and CYP2R1 polymorphisms. Diabetes Obes. Metab. 2008, 10, 683–685.
- Haussler, M.R.; Jurutka, P.W.; Mizwicki, M.; Norman, A.W. Vitamin D receptor (VDR)-mediated actions of 1α,25(OH)2vitamin D3: Genomic and non-genomic mechanisms. Best Pract. Res. Clin. Endocrinol. Metab. 2011, 25, 543–559.
- Triantos, C.; Aggeletopoulou, I.; Kalafateli, M.; Spantidea, P.I.; Vourli, G.; Diamantopoulou, G.; Tapratzi, D.; Michalaki, M.; Manolakopoulos, S.; Gogos, C.; et al. Prognostic significance of vitamin D receptor (VDR) gene polymorphisms in liver cirrhosis. Sci. Rep. 2018, 8, 14065.
- Charoenngam, N.; Holick, M.F. Immunologic Effects of Vitamin D on Human Health and Disease. Nutrients 2020, 12, 2097.
- Zhang, X.; Zhou, M.; Guo, Y.; Song, Z.; Liu, B. 1,25-Dihydroxyvitamin D3 Promotes High Glucose-Induced M1 Macrophage Switching to M2 via the VDR-PPARγ Signaling Pathway. Biomed. Res. Int. 2015, 2015, 157834.
- Powe, C.E.; Evans, M.K.; Wenger, J.; Zonderman, A.B.; Berg, A.H.; Nalls, M.; Tamez, H.; Zhang, D.; Bhan, I.; Karumanchi, S.A.; et al. Vitamin D–Binding Protein and Vitamin D Status of Black Americans and White Americans. N. Engl. J. Med. 2013, 369, 1991–2000.
- Witke, W.F.; Gibbs, P.E.M.; Zielinski, R.; Yang, F.; Bowman, B.H.; Dugaiczyk, A. Complete Structure of the Human Gc Gene: Differences and Similarities between Members of the Albumin Gene Family. Genomics 1993, 16, 751–754.
- Ahn, J.; Yu, K.; Stolzenberg-Solomon, R.; Simon, K.C.; McCullough, M.L.; Gallicchio, L.; Jacobs, E.J.; Ascherio, A.; Helzlsouer, K.; Jacobs, K.B.; et al. Genome-wide association study of circulating vitamin D levels. Hum. Mol. Genet. 2010, 19, 2739–2745.
- Thanapirom, K.; Suksawatamnuay, S.; Sukeepaisarnjareon, W.; Tanwandee, T.; Charatcharoenwitthaya, P.; Thongsawat, S.; Leerapun, A.; Piratvisuth, T.; Boonsirichan, R.; Bunchorntavakul, C.; et al. Genetic variation in the vitamin D pathway CYP2R1 gene predicts sustained HBeAg seroconversion in chronic hepatitis B patients treated with pegylated interferon: A multicenter study. PLoS ONE 2017, 12, e0173263.
- Wang, T.J.; Zhang, F.; Richards, J.B.; Kestenbaum, B.; van Meurs, J.B.; Berry, D.; Kiel, D.P.; Streeten, E.A.; Ohlsson, C.; Koller, D.L.; et al. Common genetic determinants of vitamin D insufficiency: A genome-wide association study. Lancet 2010, 376, 180–188.
- Laursen, J.H.; Søndergaard, H.B.; Albrechtsen, A.; Frikke-Schmidt, R.; Koch-Henriksen, N.; Sørensen, P.S.; Sellebjerg, F.; Oturai, A. Genetic and environmental determinants of 25-hydroxyvitamin D levels in multiple sclerosis. Mult. Scler. J. 2015, 21, 1414–1422.
- Yan, X.; Zhao, Y.; Pan, J.; Fang, K.; Wang, Y.; Li, Z.; Chang, X. Vitamin D-binding protein (group-specific component) has decreased expression in rheumatoid arthritis. Clin. Exp. Rheumatol. 2012, 30, 525–533.
- Yoshida, S.; Ikari, K.; Furuya, T.; Toyama, Y.; Taniguchi, A.; Yamanaka, H.; Momohara, S. A GC polymorphism associated with serum 25-hydroxyvitamin D level is a risk factor for hip fracture in Japanese patients with rheumatoid arthritis: 10-year follow-up of the Institute of Rheumatology, Rheumatoid Arthritis cohort study. Arthritis Res. Ther. 2014, 16, R75.
- Bae, S.-C.; Lee, Y.H. Vitamin D level and risk of systemic lupus erythematosus and rheumatoid arthritis: A Mendelian randomization. Clin. Rheumatol. 2018, 37, 2415–2421.
- Zhu, J.; DeLuca, H.F. Vitamin D 25-hydroxylase—Four decades of searching, are we there yet? Arch. Biochem. Biophys. 2012, 523, 30–36.
- Jeon, S.-M.; Shin, E.-A. Exploring vitamin D metabolism and function in cancer. Exp. Mol. Med. 2018, 50, 20.
- Falleti, E.; Cmet, S.; Fabris, C.; Fattovich, G.; Cussigh, A.; Bitetto, D.; Ceriani, E.; Lenisa, I.; Dissegna, D.; Ieluzzi, D.; et al. Genetic Polymorphisms of Vitamin D Pathway Predict Antiviral Treatment Outcome in Slow Responder Naïve Patients with Chronic Hepatitis C. PLoS ONE 2013, 8, e80764.
- Maodobra, M. The Role of Single Nucleotide Polymorphisms of Untranslated Regions (Utrs) in Insulin Resistance Pathogenesis in Patients with Type 2 Diabetes. In Medical Complications of Type 2 Diabetes; Croniger, C., Ed.; IntechOpen: London, UK, 2011.
- Hewison, M.; Zehnder, D.; Bland, R.; Stewart, P. 1alpha-Hydroxylase and the action of vitamin D. J. Mol. Endocrinol. 2000, 25, 141–148.
- Lange, C.M.; Bojunga, J.; Ramos-Lopez, E.; Von Wagner, M.; Hassler, A.; Vermehren, J.; Herrmann, E.; Badenhoop, K.; Zeuzem, S.; Sarrazin, C. Vitamin D deficiency and a CYP27B1-1260 promoter polymorphism are associated with chronic hepatitis C and poor response to interferon-alfa based therapy. J. Hepatol. 2011, 54, 887–893.
- Sundqvist, E.; Bäärnhielm, M.; Alfredsson, L.; Hillert, J.; Olsson, T.; Kockum, I. Confirmation of association between multiple sclerosis and CYP27B1. Eur. J. Hum. Genet. 2010, 18, 1349–1352.
- Hyppönen, E.; Berry, D.J.; Wjst, M.; Power, C. Serum 25-hydroxyvitamin D and IgE—A significant but nonlinear relationship. Allergy 2009, 64, 613–620.
- Gado, K.H.; Gado, T.H.; Samie, R.M.A.; Khalil, N.M.; Emam, S.L.; Fouad, H.H. Clinical significance of vitamin D deficiency and receptor gene polymorphism in systemic lupus erythematosus patients. Egypt. Rheumatol. 2017, 39, 159–164.
- Zenata, O.; Vrzal, R. Fine tuning of vitamin D receptor (VDR) activity by post-transcriptional and post-translational modifications. Oncotarget 2017, 8, 35390–35402.
- Sunn, K.L.; Cock, T.-A.; Crofts, L.A.; Eisman, J.A.; Gardiner, E.M. Novel N-Terminal Variant of Human VDR. J. Mol. Endocrinol. 2001, 15, 1599–1609.
- Hasan, H.A.; AbuOdeh, R.O.; Muda, W.A.M.B.W.; Mohamed, H.J.B.J.; Samsudin, A.R. Association of Vitamin D receptor gene polymorphisms with metabolic syndrome and its components among adult Arabs from the United Arab Emirates. Diabetes Metab. Syndr. 2017, 11, S531–S537.
- Arai, H.; Miyamoto, K.-I.; Taketani, Y.; Yamamoto, H.; Iemori, Y.; Morita, K.; Tonai, T.; Nishisho, T.; Mori, S.; Takeda, E. A Vitamin D Receptor Gene Polymorphism in the Translation Initiation Codon: Effect on Protein Activity and Relation to Bone Mineral Density in Japanese Women. J. Bone Miner. Res. 1997, 12, 915–921.
- Al-Nahas, Z. 25-hydroxyvitamin D3 deficiency and vitamin D receptor polymorphisms in Egyptian patients with Behçet’s disease: A pilot study. Clin. Rheumatol. 2017, 12, 20–27.
- Uitterlinden, A.G.; Fang, Y.; van Meurs, J.B.J.; Pols, H.A.P.; van Leeuwen, J.P.T.M. Genetics and biology of vitamin D receptor polymorphisms. Gene 2004, 338, 143–156.
- Gross, C.; Krishnan, A.V.; Malloy, P.J.; Eccleshall, T.R.; Zhao, X.-Y.; Feldman, D. The Vitamin D Receptor Gene Start Codon Polymorphism: A Functional Analysis of FokI Variants. J. Bone Miner. Res. 1998, 13, 1691–1699.
- Mahto, H.; Tripathy, R.; Das, B.K.; Panda, A.K. Association between vitamin D receptor polymorphisms and systemic lupus erythematosus in an Indian cohort. Int. J. Rheum. Dis. 2018, 21, 468–476.
- Yiallourou, A.I.; Ekonomou, E.; Tsamadias, V.; Nastos, K.; Karapanos, K.; Papaconstantinou, I.; Theodosopoulos, T.; Contis, J.; Papalambros, E.; Voros, D.; et al. Association of FokI and PvuII polymorphisms with breast cancer staging and survival among Caucasian women: A prospective study. J. BU ON Off. J. Balk. Union Oncol. 2014, 19, 633–642.
- Li, L.; Wu, B.; Liu, J.-Y.; Yang, L.-B. Vitamin D Receptor Gene Polymorphisms and Type 2 Diabetes: A Meta-analysis. Arch. Med. Res. 2013, 44, 235–241.
- Goswami, R. Primer on the metabolic bone diseases and disorders of mineral metabolism. Indian J. Med. Res. 2016, 144, 489–490.
- Garavito, G.; Egea, E.; Fang, L.; Malagón, C.; Olmos, C.; González, L.; Guarnizo, P.; Aroca, G.; López, G.; Iglesias, A. Association of polymorphic variants of PTPN22, TNF and VDR systems in children with lupus nephritis: A study in trios of Colombian families. Biomedica 2017, 37, 260–266, doi:10.7705/biomedica.v37i3.3247.
- Salimi, S.; Eskandari, F.; Rezaei, M.; Sandoughi, M. Vitamin D Receptor rs2228570 and rs731236 Polymorphisms are Susceptible Factors for Systemic Lupus Erythematosus. Adv. Biomed. Res. 2019, 8, 48.
- López-Mejías, R.; Genre, F.; Remuzgo-Martínez, S.; Robledo, G.; Llorca, J.; Corrales, A.; González-Juanatey, C.; Ubilla, B.; Mijares, V.; Pina, T.; et al. Vitamin D receptor GATG haplotype association with atherosclerotic disease in patients with rheumatoid arthritis. Atherosclerosis 2016, 245, 139–142.
- Ghaly, M.S.; Badra, D.I.; Dessouki, O.; Elmaraghy, N.N.; Hassan, R. Vitamin D receptor Fok1 & Bsm 1 Gene Polymorphisms in Systemic Lupus Erythematosus and Osteoarthritis: Autoimmune Inflammatory versus Degenerative Model. Egypt. J. Immunol. 2017, 24, 151–164.
- Babenko, S.A.; Alifirova, V.M.; Orlova, I.; Puzyrev, V.P. [The VDR gene polymorphism in patients with multiple sclerosis]. Zhurnal Nevrologii i Psikhiatrii Imeni S.S. Korsakova 2009, 109 (Suppl. 2), 23–27.
