Glutamate decarboxylase (l-glutamate-1-carboxylase, GAD; EC 4.1.1.15) is a pyridoxal-5’-phosphate-dependent enzyme that catalyzes the irreversible α-decarboxylation of l-glutamic acid to γ-aminobutyric acid (GABA) and CO2. The enzyme is widely distributed in eukaryotes as well as prokaryotes, where it—together with its reaction product GABA—fulfils very different physiological functions. The occurrence of gad genes encoding GAD has been shown for many microorganisms, and GABA-producing lactic acid bacteria (LAB) have been a focus of research during recent years. A wide range of traditional foods produced by fermentation based on LAB offer the potential of providing new functional food products enriched with GABA that may offer certain health-benefits. Different GAD enzymes and genes from several strains of LAB have been isolated and characterized recently.
- Glutamate Decarboxylase
- lactic acid bacteria
- amino acids
1. Introduction
Glutamate decarboxylase is an intracellular enzyme that is found ubiquitously in eukaryotes and prokaryotes. GAD exhibits different physiological roles, especially in vertebrates and plants, and its presence is highly variable among organisms [52][1]. GAD is a PLP-dependent enzyme and as such belongs to the PLP-dependent enzyme superfamily. This superfamily comprises seven different folds [67][2] with GAD from LAB showing the type-I fold of PLP-dependent enzymes [68][3]. A number of important catalytic reactions including α- and β-eliminations, decarboxylation, transamination, racemization, and aldol cleavage are catalyzed by various members of this superfamily of enzymes [69][4]. GAD activity relies on the binding of its co-factor PLP, and belongs to group II of PLP-dependent decarboxylases [70][5]. In GAD from L. brevis CGMCC 1306, the active site entrance is located at the re-face of the cofactor PLP. PLP is covalently attached to a lysine (K279) via an imine linkage (Figure 1), referred to as an internal aldimine [68,71][3][6]. This lysine is strictly conserved in group II PLP-dependent decarboxylases. The corresponding lysine in E. coli GAD is at position 276, and when mutating this residue, the variant has less flexibility and affinity to both its substrate and the cofactor [72][7]. In addition to this covalent attachment, PLP is positioned in the active site via a number of H bonds between the phosphate group of PLP and surrounding amino acids, while the pyridine ring of PLP forms hydrophobic interactions with side chains of various amino acids in the active site [68][3].
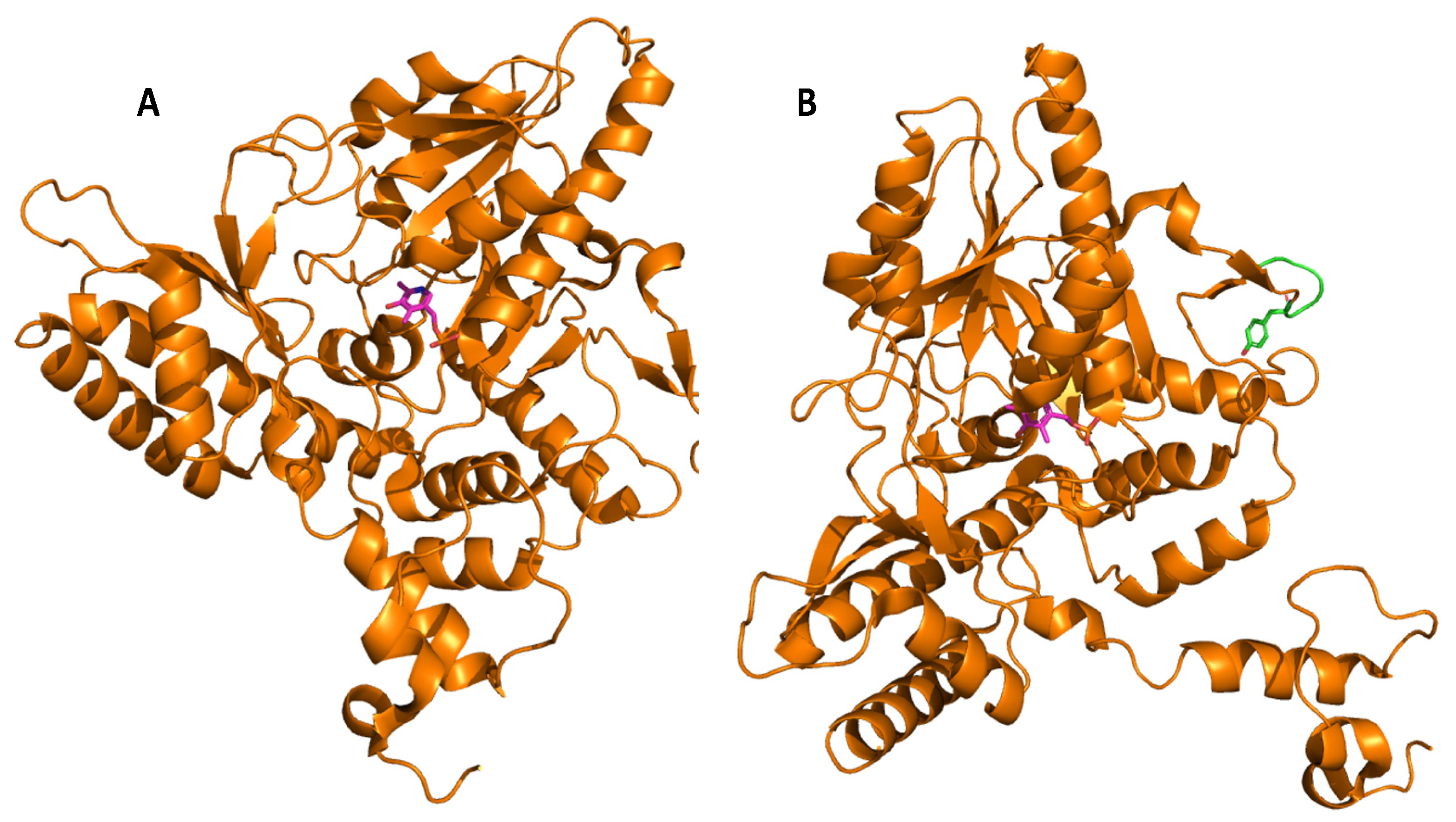
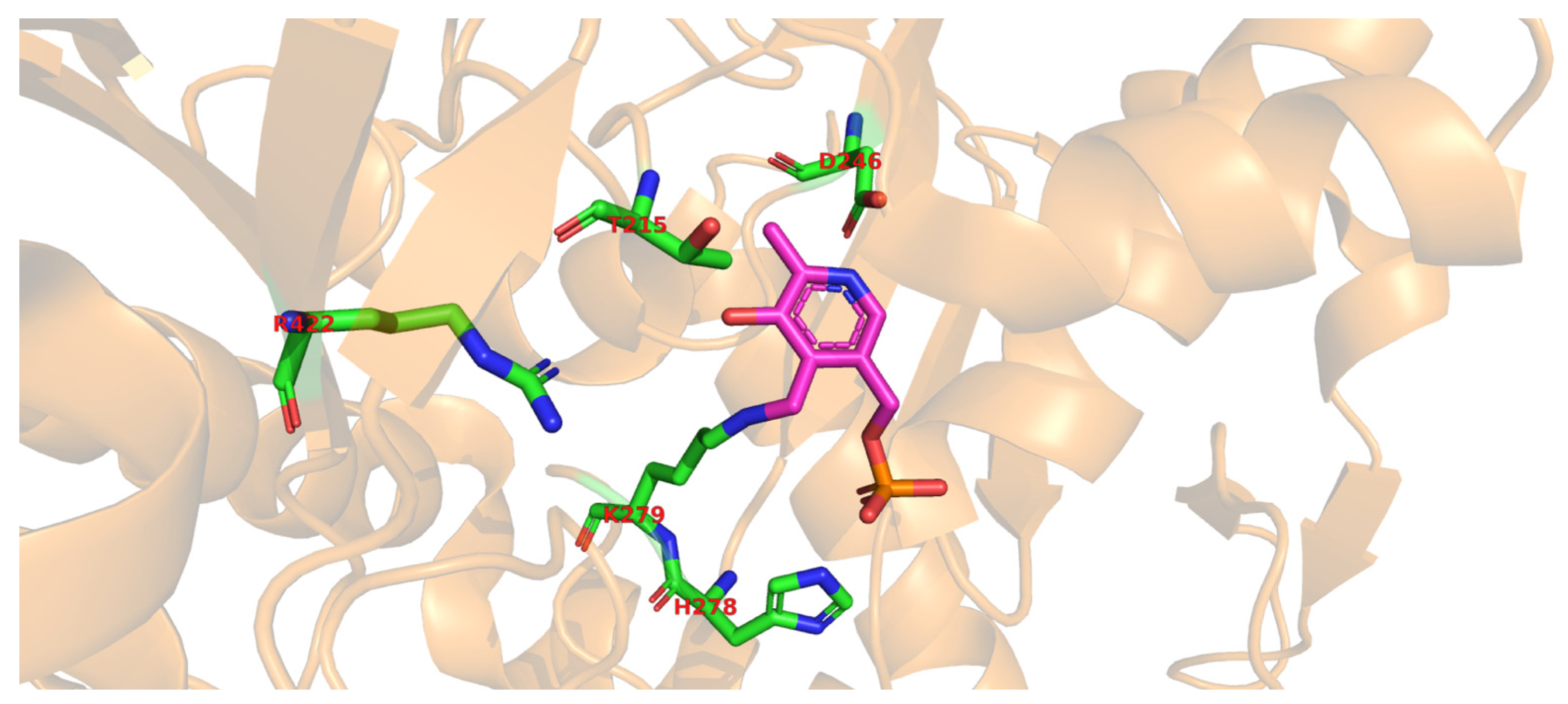
Molecular docking of the substrate glutamate into the active-site of the holo-form of L. brevis GAD showed several noncovalent interactions including hydrogen bonds between the O2, the O3 and the O4 atoms of the substrate L-Glu to various parts of the GAD polypeptide chain. Furthermore, electrostatic interactions between the negatively charged oxygen atom of the α-carboxyl and the γ-carboxyl group of L-Glu and the positively charged nitrogen atom of residue R422 as well as H278 and K279 (Figure 2), respectively, were proposed [68][3]. The flexible loop residue Tyr308-Glu312 in L. brevis GAD is located near the substrate-binding site (Figure 1). This loop is important for the catalytic reaction, and the conserved residue Tyr308 plays a crucial role in decarboxylation of L-Glu. Thr 215 and Asp246 are the two catalytic residues in L. brevis GAD (Figure 2), which are also highly conserved and promote decarboxylation of L-Glu [68,71,73][3][6][8].
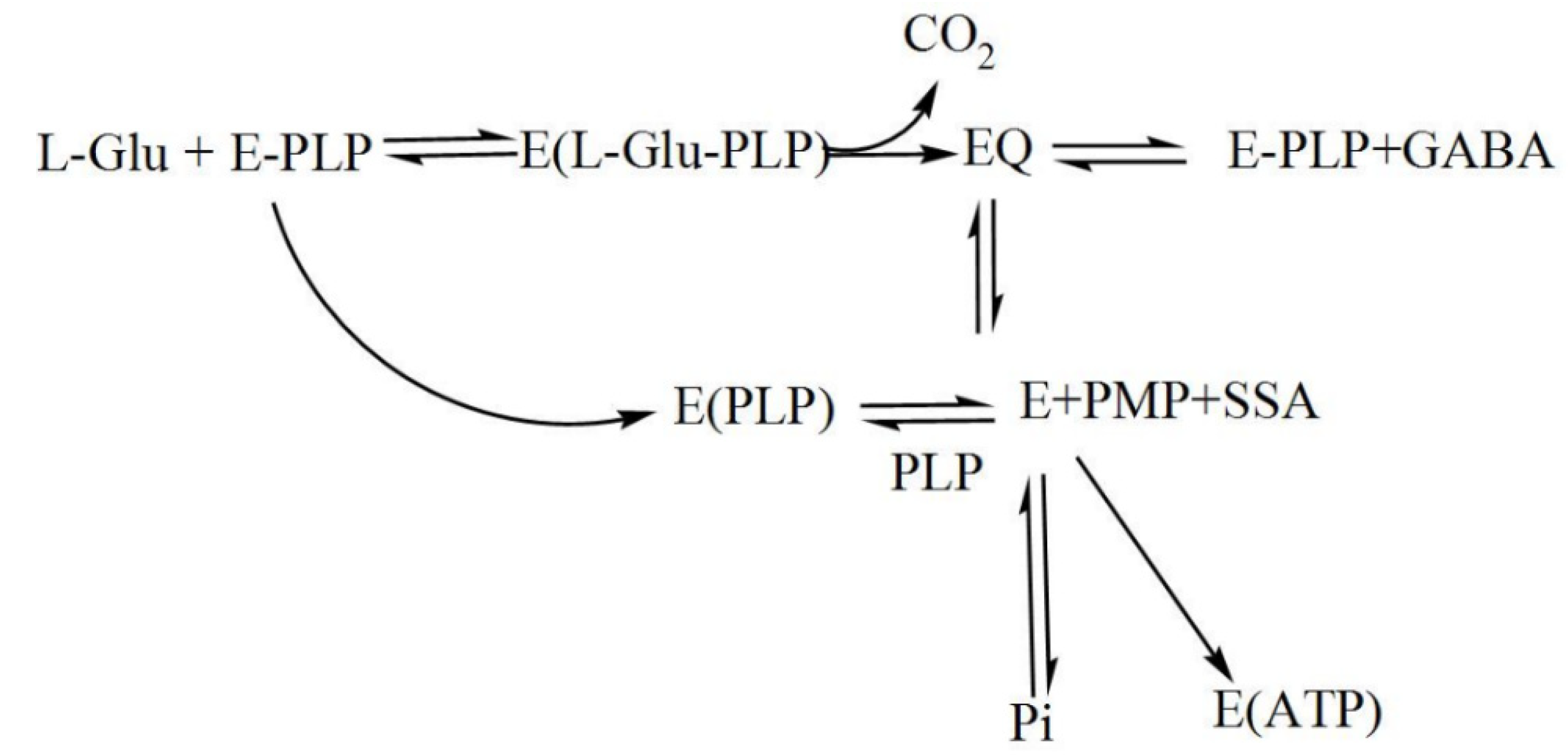
During catalysis, a transamination reaction occurs, and PLP, which is covalently attached to a Lys in the active site of GAD in its resting state, now becomes covalently bonded to the substrate glutamate, forming a Schiff base or what is referred to as an external aldimine. This Schiff base can then be transformed to a quinonoid intermediate [67,74][2][9]. In a small fraction of catalytic cycles, when glutamate is decarboxylated, a subsequent alternative transamination of the quinonoid intermediate of the reaction can occur, and succinic semialdehyde (SSA) and pyridoxamine-5′-phosphate (PMP) are formed. The latter will immediately be released from the enzyme, resulting in inactive apoGAD (Figure 3), which can be regenerated to the active GAD–PLP complex when free pyridoxal-5′-phosphate is present, thus completing a cycle of inactivation and activation. However, when free PLP is not present, GAD will be inactivated as a function of time and substrate concentration [62,67,68,69,74,75,76,77].[10][2][3][9][11][12][13]
2. The Role of Glutamate Decarboxylase in the Manufacturing of Bio-Based Industrial Chemicals
Agricultural waste and waste streams from biofuel production are now being considered as a low-cost source of glutamate for biotechnological conversion into GABA and production of bio-based chemicals [107][14]. These protein-rich materials are mainly bioethanol by-product streams including dried distiller’s grains with solubles (DDGS) from maize and wheat, or vinasse from sugarcane or sugar beet, but also plant leaves, oil, or biodiesel by-products and slaughterhouse waste. In the future, algae could also provide an additional source for biodiesel and thus become a natural low-cost source of glutamic acid.
The protein-rich fraction of plants can be further split into more- and less-nutritious fractions, for example, by hydrolyzing the proteins and separating the essential (nutritious) amino acids from the non-essential (less nutritious) ones. Non-essential amino acids such as glutamic acid and aspartic acid, which have no significant value in animal feed, can be utilized for preparing functionalized chemicals. Recently, a by-product from the tuna canning industry, tuna condensate, was shown to be a useful material for the production of GABA. Tuna condensate contains significant amounts of glutamine, but relatively little glutamate. Glutamine was first converted to glutamate by a glutaminase from Candida rugosa, and in a second step, L. futsaii GAD converted glutamate to GABA. Both steps were catalyzed by immobilized whole cells [108][15]. Recently, it was shown that supplementation of arginine to media containing glutamate could enhance GABA production, and that the simultaneous addition of arginine, malate, and glutamate enabled GABA production already during exponential growth at relatively high pH (6.5) [109][16].
The structure of glutamic acid resembles many industrial intermediates, so it can be transformed into a variety of chemicals using a relatively limited number of steps. Decarboxylation of glutamic acid to GABA, enzymatically performed by GAD, is an important reaction of the pathway from glutamic acid to a range of molecules. GABA is, for example, an intermediate for the synthesis of pyrrolidones. Such an approach can be used to produce N-methyl-2-pyrrolidone (NMP), which is used as an industrial solvent. Combining the enzymatic decarboxylation of glutamate performed by GAD with the one-pot cyclization of GABA to 2-pyrrolidone and subsequent methylation will thus yield NMP [110][17]. Another interesting material synthesized by ring-opening polymerization of 2-pyrrolidone is Nylon 4 [111][18], a four-carbon polyamide suitable for application as an engineering plastic due to its superior thermal and mechanical properties [112][19]. Contrary to other nylon polymers, Nylon 4 is heat-resistant, biodegradable, biocompatible, and compostable [112][19].
References
- Ueno, H. Enzymatic and structural aspects on glutamate decarboxylase. J. Mol. Catal. B Enzym. 2000, 10, 67–79.
- Steffen-Munsberg, F.; Vickers, C.; Kohls, H.; Land, H.; Mallin, H.; Nobili, A.; Skalden, L.; Bergh, T.V.D.; Joosten, H.-J.; Berglund, P.; et al. Bioinformatic analysis of a PLP-dependent enzyme superfamily suitable for biocatalytic applications. Biotechnol. Adv. 2015, 33, 566–604.
- Huang, J.; Fang, H.; Gai, Z.-C.; Mei, J.-Q.; Li, J.-N.; Hu, S.; Lv, C.-J.; Zhao, W.-R.; Mei, L.-H. Lactobacillus brevis CGMCC 1306 glutamate decarboxylase: Crystal structure and functional analysis. Biochem. Biophys. Res. Commun. 2018, 503, 1703–1709.
- Rocha, J.F.; Pina, A.F.; Sousa, S.F.; Cerqueira, N.M.F.S.A. PLP-dependent enzymes as important biocatalysts for the pharmaceutical, chemical and food industries: A structural and mechanistic perspective. Catal. Sci. Technol. 2019, 9, 4864–4876.
- Sandmeier, E.; Hale, T.I.; Christen, P. Multiple evolutionary origin of pyridoxal-5′-phosphate-dependent amino acid decarboxylases. Eur. J. Biochem. 1994, 221, 997–1002.
- Yu, K.; Lin, L.; Hu, S.; Huang, J.; Mei, L. C-terminal truncation of glutamate decarboxylase from Lactobacillus brevis CGMCC 1306 extends its activity toward near-neutral pH. Enzym. Microb. Technol. 2012, 50, 263–269.
- Tramonti, A.; John, R.A.; Bossa, F.; De Biase, D. Contribution of Lys276 to the conformational flexibility of the active site of glutamate decarboxylase from Escherichia coli. Eur. J. Biochem. 2002, 269, 4913–4920.
- Kim, S.-H.; Shin, B.-H.; Kim, Y.-H.; Nam, S.-W.; Jeon, S.-J. Cloning and expression of a full-length glutamate decarboxylase gene from Lactobacillus brevis BH2. Biotechnol. Bioprocess Eng. 2007, 12, 707–712.
- Wang, C.; Zhu, R.; Sun, H.; Li, B. Quantum chemistry studies of the catalysis mechanism differences between the two isoforms of glutamic acid decarboxylase. J. Mol. Model. 2013, 19, 705–714.
- Yunes, R.A.; Poluektova, E.U.; Dyachkova, M.S.; Klimina, K.M.; Kovtun, A.S.; Averina, O.V.; Orlova, V.S.; Danilenko, V.N. GABA production and structure of gadB/gadC genes in Lactobacillus and Bifidobacterium strains from human microbiota. Anaerobe 2016, 42, 197–204.
- Chang, C.; Zhang, J.; Ma, S.; Wang, L.; Wang, D.; Zhang, J.; Gao, Q. Purification and characterization of glutamate decarboxylase from Enterococcus raffinosus TCCC11660. J. Ind. Microbiol. Biotechnol. 2017, 44, 817–824.
- Battaglioli, G.; Liu, H.; Martin, D.L. Kinetic differences between the isoforms of glutamate decarboxylase: Implications for the regulation of GABA synthesis. Neurochemistry 2003, 86, 879–887.
- Jansonius, J.N. Structure, evolution and action of vitamin B6-dependent enzymes. Curr. Opin. Struct. Biol. 1998, 8, 759–769.
- Lammens, T.M.; Franssen, M.C.R.; Scott, E.L.; Sanders, J.P.M. Availability of protein-derived amino acids as feedstock for the production of bio-based chemicals. Biomass Bioenergy 2012, 44, 168–181.
- Sanchart, C.; Watthanasakphuban, N.; Boonseng, O.; Nguyen, T.-H.; Haltrich, D.; Maneerat, S. Tuna condensate as a promising low-cost substrate for glutamic acid and GABA formation using Candida rugosa and Lactobacillus futsaii. Process. Biochem. 2018, 70, 29–35.
- Laroute, V.; Yasaro, C.; Narin, W.; Mazzoli, R.; Pessione, E.; Cocaign-Bousquet, M.; Loubière, P. GABA production in Lactococcus lactis is enhanced by arginine and co-addition of malate. Front. Microbiol. 2016, 7, 1050.
- Lammens, T.M.; Franssen, M.C.R.; Scott, E.L.; Sanders, J.P.M. Synthesis of biobased N-methylpyrrolidone by one-pot cyclization and methylation of γ-aminobutyric acid. Green Chem. 2010, 12, 1430–1436.
- Park, S.J.; Kim, E.Y.; Noh, W.; Oh, Y.H.; Kim, H.Y.; Song, B.K.; Cho, K.M.; Hong, S.H.; Lee, S.H.; Jegal, J. Synthesis of nylon 4 from γ-aminobutyrate (GABA) produced by recombinant Escherichia coli. Bioprocess Biosyst. Eng. 2013, 36, 885–892.
- Tokiwa, Y.; Calabia, B.P.; Ugwu, C.U.; Aiba, S. Biodegradability of plastics. Int. J. Mol. Sci. 2009, 10, 3722–3742.
