1. Background of Cell Proteomic Footprinting
Many molecules within a cell are associated with basic cellular functions such as biosynthesis, energy balance, and cell division, and the antigenic determinants associated with the molecules involved are mostly common to all cells in the body
[1][2][3]. The surface of the cell is the place where its individuality is most expressed. A variety of environmental factors, including contact with neighboring cells, contact with surrounding structures, and the presence of specific receptors, contribute to the unique ‘face’ of each cell type
[4][5][6]. A clear example of this is that when cancer cells appear in the body, the immune system recognizes and removes them, even if the morphology of the cancer cells is close to normal cells. The antibodies and killer cells sent by the immune system only recognize the antigens that are present on the surface of cancer cells, as they do not penetrate inside a living cell. This fact demonstrates that the cell surface is sufficient for the accurate recognition of cells and their precise differentiation from each other
[5].
Therefore, to develop immunotherapeutic agents that are effective against cancer cells, such as cancer vaccines, it is necessary to ‘read’ the ‘face’ of cancer target cells to ensure that the vaccine has the complimentary antigen composition. The bulk of specific molecules on the cell surface are proteins and, associated with them, carbohydrates. To ‘read’ the cell surface, it must first be isolated. With this, it is necessary to treat living cells with a protease, e.g., trypsin. Since the protease is quite large molecule, it does not penetrate the cell and therefore only cleaves the proteins that are present on the surface
[7][8].
This creates the basis of the cellular proteomic footprinting (CPF). Trypsin occupies a central role in proteomics—an omics science and a technology platform used for the high-throughput analysis of proteins in biological samples
[9][10]. The occurrence of these sites in proteins is such that the number and size (molecular weight) of cleaved fragments is sufficient to obtain a highly protein-specific characteristic (known as peptide fingerprint
[11]).
Proteomics has evolved over time, resulting in perfected proteome analysis protocols, the availability of required pure proteomics-grade reagents (including proteomics-grade trypsin), and experience in proteomic data processing
[12]. All of this established the background for the emergence of CPF
[13], which is essentially the use of trypsin to treat the surface of cells to obtain a specific proteomic pattern consisting of trypsinolytic fragments of cell surface proteins. Since this pattern is specific to cells, it enables them to be identified or authenticated, confirming that they have expected therapeutic properties.
CPF is largely relevant in the development and manufacture of whole-cell, cell lysate, cell membrane, and antigenic essence-based vaccines that utilize cancer cells and cells from the tumor microenvironment, such as microvascular endothelial cells and fibroblasts. CPF has the greatest complementarity with the technology for developing a variety of anticancer vaccines based on the
antigenic essence of cells
[14][15].
2. Cell Proteomic Footprinting
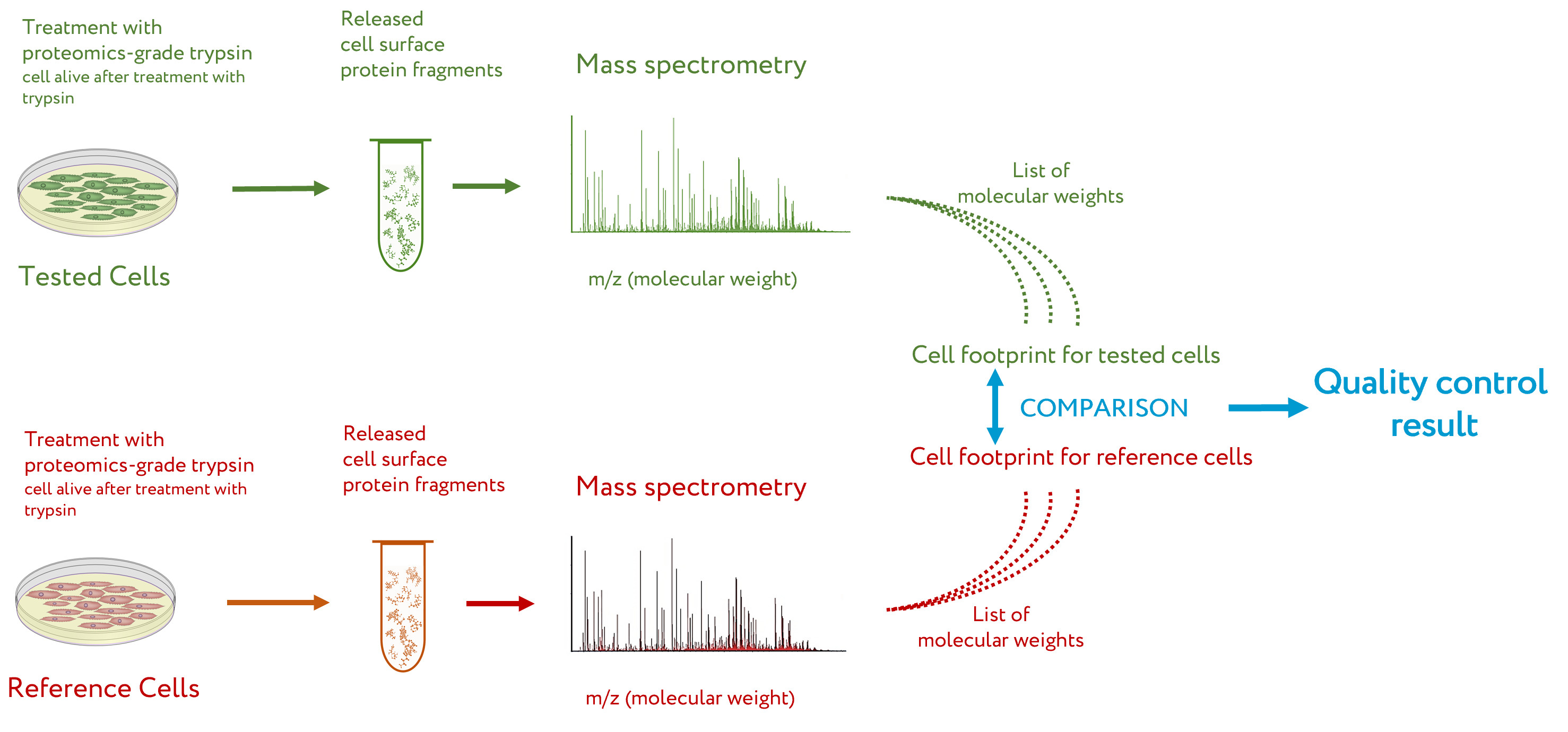
The general workflow for CPF is presented in
Figure 1 [14]. Briefly, the
cell culture to be tested by CPF is washed several times to remove traces of the culture medium, especially the proteins that can be absorbed on the cell surface. After this, the cells are treated with a solution of proteomics-grade trypsin
[16] under mild conditions, so that the cells after treatment are alive but extracellular fragments of proteins are cleaved off
[8]. The solution with cleaved protein fragments is taken, and a sample of it is subjected to peptide mass spectrometry. The obtained list of peptide masses is a cell proteomic footprint that is specific to the tested cells. By comparing this footprint with a reference footprint corresponding to the cells with the desired properties, the presence of these properties, their absence, or the degree of deviation from them is evaluated for the tested cells
[14].
Figure 1.
Workflow of cell proteomic footprinting. Adapted from
.
3. Cell Authentication by Cell Proteomic Footprinting
3.1 Cell type authentication
Cross-contamination and misidentification are common problems with cultured cells
[17][18][19][20][21][22]. It was shown that up to 36% of cell lines appear to have a different origin than their initial cell lines
[21][23]. Moreover, even the most popular cell lines, e.g. HepG2
[24], which are widely used in science, raises questions due to their heterogeneity. Today, a method of cell authentication based on relatively stable genotype data, short tandem repeat (STR) markers (tiny repetitive segments of DNA found between genes), is widely used
[25][26][27][28][29].
However, for cell products, it is important to not only authenticate the cell type at the genome level. For example, an established cell culture can multiply many times and exhibit characteristic properties, including therapeutic ones. However, close to senescence, the primary culture degenerates and begins to die
[30][31][32][33][34], while the genotype (both at the beginning and at senescence) is the same. Therefore, authentication at the molecular phenotype level (post-genome level) is also needed for the development of cell products. Thus, CPF, relating to the molecular phenotype associated with the confirmation of the therapeutic qualities of cells, complements genotyping via the STR assay.
It is worth noting that subjecting trypsinolytic protein fragments to mass spectrometry for cell identification is not new and is already used in diagnostic bacteriology
[35][36][37] . Thus, the Biotyper (Bruker Daltonik, USA)
[38] identifies germs via the peptide fingerprinting of proteins
[39][40][41]. The use of CPF as a cell authentication tool is similar to that of Biotyper, but mainly intended for mammalian cells.
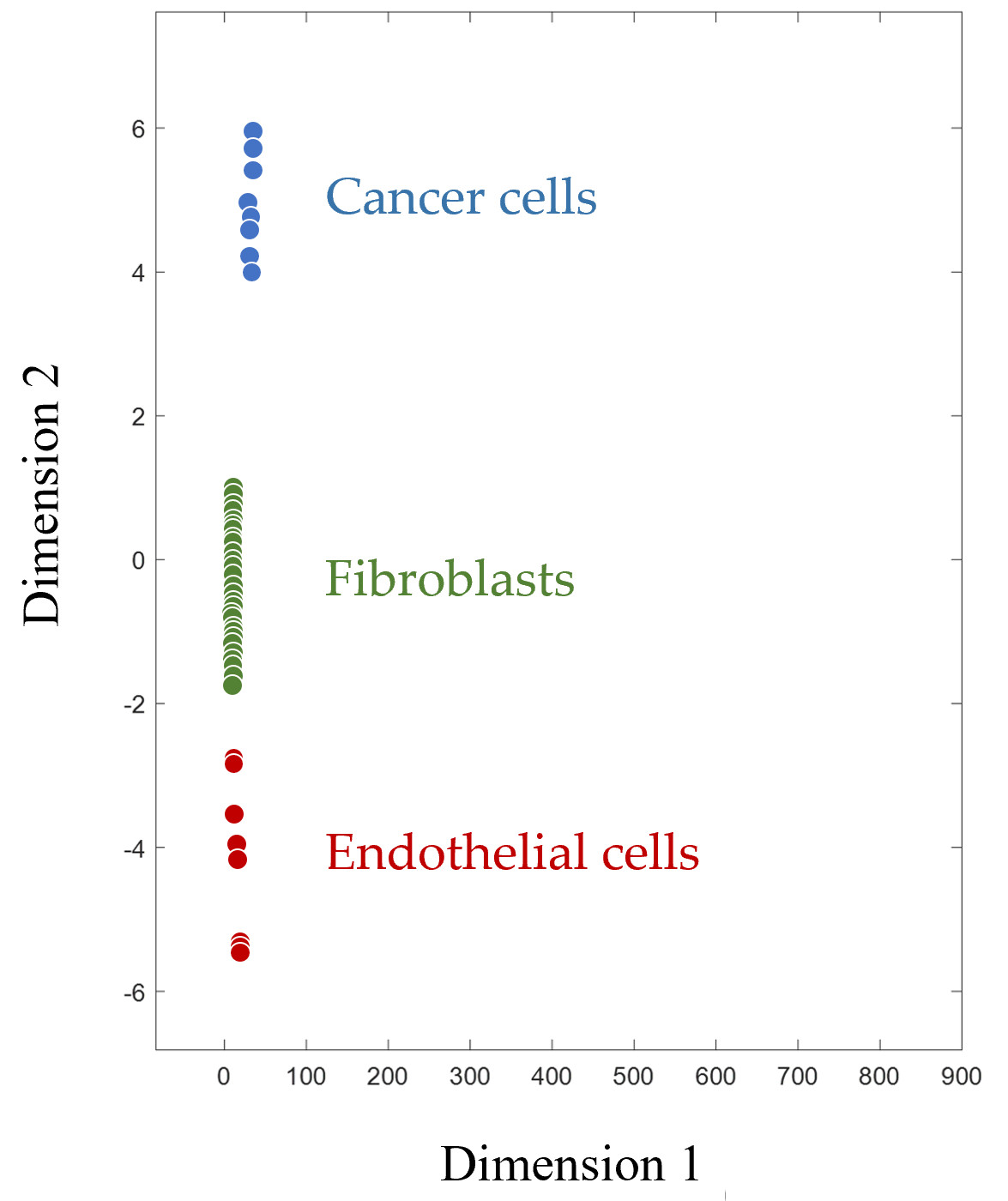
Figure 2 shows how CPF clearly divides mammalian cells into groups according to their type. Since the footprint is a multivariable characteristic, multidimensional scaling is applied for the visualization of footprints in a two-dimensional plane. Along with human cancer cells, endothelial cells (human microvascular endothelial cells, HMECs), and fibroblasts of various origins are represented. These cell types, like cancer cells themselves, are also considered targets for immunotherapy, as they are part of the tumor microenvironment and are required to ensure its functioning
[42][43][44][45][46].
Figure 2. Multidimensionally scaled cell proteomic footprints of various cells. Each point on the plot corresponds to a separate cell culture (primary culture of human microvascular endothelial cells and fibroblasts, as well as various cancer cell lines).
The proof that CPF can objectively identify and authenticate cell types with high accuracy is presented in Table 1.
Table 1.
The separation of different cell types based on CPF.
|
Separated cells
|
Separation efficacy |
The separation efficiency of different cell subpopulations based on CPF.
|
Separated subpopulations of cells
|
Separation efficacy |
| Sensitivity (%) |
Specificity (%) |
Accuracy (%) |
|---|
| Sensitivity (%) |
Specificity (%) |
Accuracy (%) |
|---|
| Cancer cells from drug-treated cancer cells |
100.0 |
90.0 |
95.0 |
| Human microvascular endothelial cells from tumor-stimulated human microvascular endothelial cells |
86.7 |
100.0 |
94.0 |
| Adipose fibroblasts from skin fibroblasts |
100.0 |
100.0 |
100.0 |
| Skin fibroblasts from dermal papilla fibroblasts |
94.4 |
66.7 |
68.8 |
| Dermal papilla fibroblasts from adipose fibroblasts |
83.3 |
83.3 |
83.3 |
| Human microvascular endothelial cells from non-endothelial cells |
100.0 |
100.0 |
100.0 |
| Cancer cells from non-cancer cells |
100.0 |
96.4 |
96.7 |
| Fibroblasts from non-fibroblasts |
98.9 |
98.8 |
98.9 |
Sensitivity—the percentage of correctly identified positive results (tested cells are correctly assigned to a specific cell type); specificity—the percentage of correctly identified negative results (tested cells correctly do not assigned to specific cell type); accuracy—the percentage of correctly identified positive and negative results. Footprints are taken from [13][47][48].
3.2 Cell subtype authentication
The cell authentication with high efficiency presented in Table 1 is more conceptual than practical. As mentioned above, the generally accepted STR-based assay copes with this task quite well. However, the authentication of subpopulations of cancer cells, as well as subpopulations of cells related to the tumor microenvironment, such as microvascular endothelial cells or fibroblasts, with a distinctive molecular phenotype and therapeutic properties that are not available for STR, relates to the direct application of CPF. Evidence that CPF can objectively identify and authenticate subpopulations of cells with different therapeutic properties is presented in Table 2.
Table 2.
Specificity—the percentage of correctly identified negative results (tested cells correctly do not assigned to specific cell type); accuracy—the percentage of correctly identified positive and negative results. Footprints are taken from [13][47][48].
3.3 Cell proteomic footprinting of fibroblast subpopulations
Fibroblast cultures are a good example to demonstrate CPF efficacy with the authentication of cell subtypes because different subtypes of fibroblasts have identical morphologies and are propagated under the same conditions. Footprints for three different subpopulations of human fibroblasts from the published studies
[13] were compared: primary cultures of dermal papilla fibroblasts, adipose-derived fibroblasts, and skin fibroblasts. These fibroblast subpopulations have identical spindle-like morphologies in cultures but exhibit different therapeutic properties according to their origin. Dermal papilla fibroblasts are trichogen cells
[49], adipose-derived fibroblasts are pluripotent
[50], and adult skin fibroblasts are better for autologous cell therapy
[51].
Table 2 demonstrates the high efficacy of separating the footprints of these fibroblast subpopulations. Thus, CPF can be recognized as a tool to authenticate cell cultures at the subpopulation level, confirming their therapeutic properties. Moreover, cancer-associated fibroblasts (CAFs) are also heterogenous population
[52] that actively involved in carcinogenesis
[53], metastasis
[54], and therapy resistance
[55]. So, CAF subpopulations represent promising targets for cancer immunotherapy
[43][44][45][56], which can also be authenticated by CPF.
3.4 Cell proteomic footprinting in cancer vaccine design
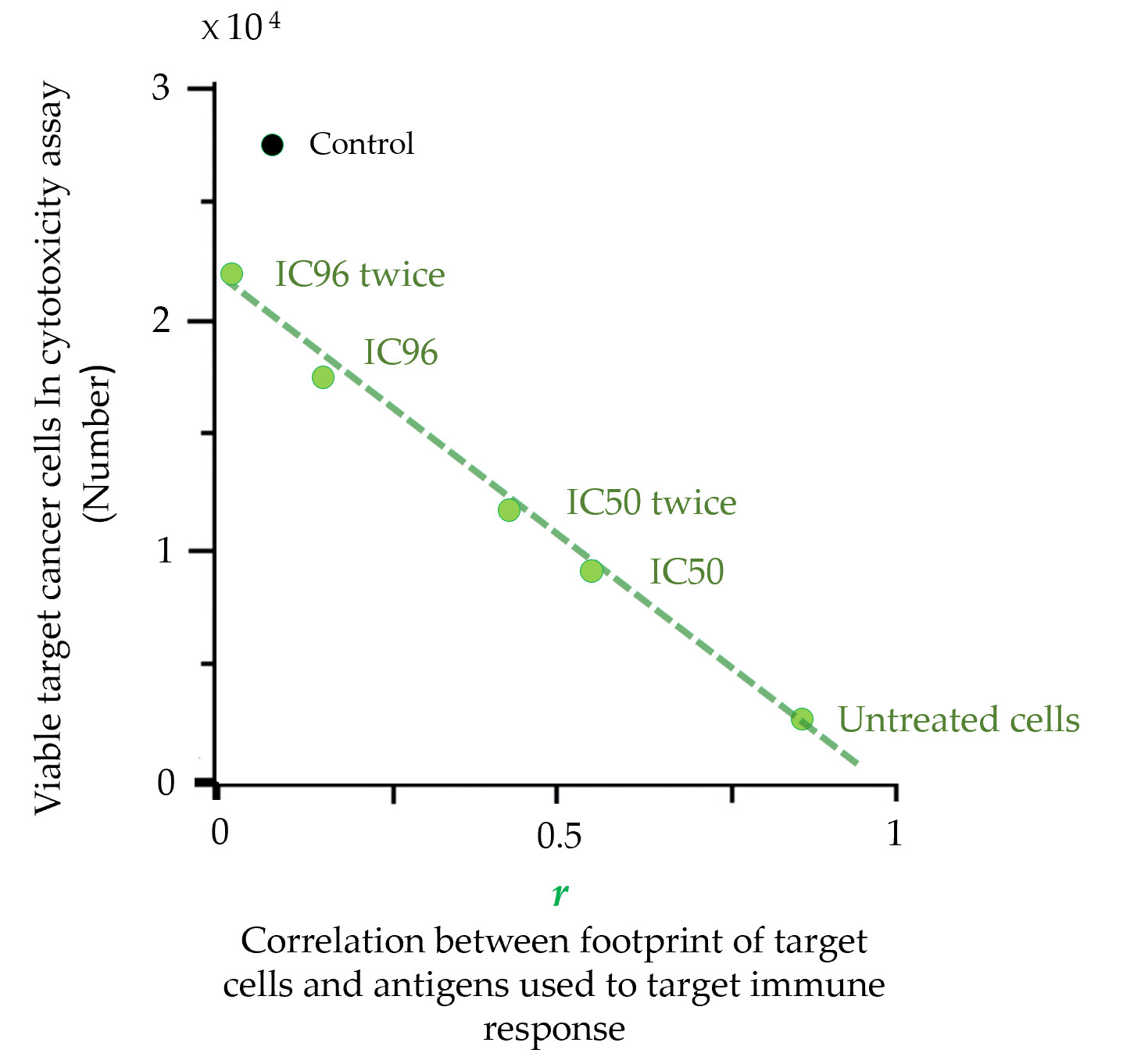
The footprint is a precise characteristic that is also directly related to the therapeutic properties of cancer cell-based vaccines. The direct connection between the footprint of cancer cells and their escape from the immune response was demonstrated in cytotoxicity assays (CTAs)
[48]. The molecular phenotype of cancer cells (MCF-7) was gradually changed by drug treatment. CTA showed that the rate of cancer cells escape from the immune response depends on the similarity between the antigen composition used to induce an immune response (
antigenic essence was used) and the proteomic footprint of targeted cancer cells (
Figure 3)
[48]. These results demonstrated that the set of antigens expressed on the surface of cancer cells can be significantly altered under drug treatment, and such alterations create cell subpopulations that completely evade the immune response. This strong argument suggests that cancer cells used to produce a cancer vaccine should undergo CPF-based quality control.
Figure 3. Escape of target cancer cells from the immune response in the cytotoxicity assay (CTA) as a result of their surface profile changes induced by the drug treatment. Points are presented for untreated MCF-7 target cells or cells treated with a single IC96 or IC50 dose of etoposide or two separate IC96 or IC50 doses (‘twice’) of etoposide (adapted from
[48]). A linear approximation is shown. Correlation coefficients (
r) were calculated for the correlation between the antigenic composition (known as cellular
antigenic essence [57]) used to induce an immune response in CTA and the proteomic footprints of target cancer cells. ‘Control’ corresponds to the immune response induced by the control antigen composition (non-relevant to target MCF-7 cells). The data in the graph show that the escape of target cells from the immune response is directly related to the degree of change in their proteomic footprint.
3.5 Cell proteomic footprinting of human microvascular endothelial cells
Endothelial cells (ECs) line the inner surface of blood vessels and constitute a selective barrier between the blood and the tissue. The importance of ECs in the context of cancer has been extensively investigated [58]. In 1945, it was reported that a tumor recruits microvasculature from surrounding tissues to support its feeding and growth [59]. This finding yielded an entire field of research that aims to inhibit new blood vessel formation [58]. Vaccination against EC in tumors offers the additional benefit that ECs are genetically more stable and therefore less likely to develop escape mutations than cancer cells [60]. Moreover, the ratio of ECs to cancer cells in tumors is approximately 1:100. The destruction of a small number of ECs can lead to vascular obstruction and arrest tumor growth because vascular integrity is essential to tumor feeding [61][62][63][64].
The molecular phenotype of ECs in the microvasculature is tissue-specific
[65][66], and if the microvasculature is involved in tumor feeding, the EC phenotype also becomes tumor-specific
[67][68][69]. This heterogeneity in the phenotype provides grounds for designing antiangiogenic vaccines for anti-cancer vaccination. The influence of tumors on the EC surface profile was investigated by culturing HMECs with a tumor-conditioned medium
[47]. CPF demonstrated that cancer cells induce HMEC subpopulations with statistically significant phenotypic differences from the initial cells (
Table 2) that are directly connected with their escape from the immune system. This finding provides a strong justification for designing and manufacturing an antiangiogenic cancer vaccine that targets tumor vessels under CPF control.