Deep learning has become a prominent approach in Alzheimer’s disease (AD) detection using medical image data, incorporating modalities like PET and MRIpositron emission tomography (PET) and magnetic resonance imaging (MRI). These advances in deep learning and multimodal imaging have improved AD detection accuracy and effectiveness, leveraging CNNs, RNNsconvolutional neural networks (CNNs), recurrent neural networks (RNNs), and generative modelling techniques.
- Alzheimer’s disease
- AD detection
- convolutional neural network
- deep learning
1. Introduction
2. Alzheimer’s Disease Detection System
Figure 1 illustrates the AD detection system, an intricate and comprehensive framework designed to facilitate the efficient detection of AD. This system harnesses the synergistic integration of essential components, including brain scans, preprocessing techniques, data management strategies, deep learning models, and evaluation. Together, these elements establish a robust foundation for the system, ensuring its effectiveness, reliability, and precision.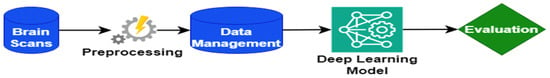
2.1. Brain Scans
2.2. Preprocessing
2.3. Data Management
2.4. Deep Learning Model
2.1. Brain Scans
2.2. Preprocessing
2.3. Data Management
2.4. Deep Learning Model
3. Deep Learning for Alzheimer’s Disease Detection
3.1. Convolutional Neural Networks for AD Detection
Convolutional neural networks (CNNs) have gained prominence in medical imaging for AD detection, as they excel at learning hierarchical features from raw image data, enabling accurate predictions [157][50]. CNNs’ promising results in AD detection have garnered attention from researchers and clinicians.
3.1.1. Neuroimaging and CNNs for AD Analysis
3.1.2. CNN-3D Architecture for AD Classification
3.1.3. GANs for Data Augmentation and Enhancement
5.1.4. Transfer Learning and Multimodal Fusion
3.1.5. Temporal Convolutional Networks (TCNs)
3.1.6. Dataset Quality and Interpretable Models
3.1.7. Convolutional Neural Network (CNN) Studies for AD Detection
Convolutional neural networks (CNNs) have demonstrated remarkable achievements in tasks such as organ segmentation [193,194,195][52][53][54] and disease detection [196,197,198][55][56][57] within the field of medical imaging. By leveraging neuroimaging data, these models can uncover hidden representations, establish connections between different components of an image, and identify patterns related to diseases [199][58]. They have been successfully applied to diverse medical imaging modalities, encompassing structural MRI [200][59], functional MRI [201,202][60][61] (fMRI), PET [203[62][63],204], and diffusion tensor imaging (DTI) [205,206,207][64][65][66]. Consequently, researchers have begun exploring the potential of deep learning models in detecting AD using medical images [61,208,209,210][67][68][69][70].
3.1.8. Performance Comparison
3.1.9. Meaningful Insights
The application of convolutional neural networks (CNNs) in Alzheimer’s disease (AD) detection has unveiled several meaningful insights. CNNs, particularly 3D architectures, have showcased their prowess in deciphering complex patterns within volumetric neuroimaging data. One remarkable insight is the ability of CNNs to extract hierarchical features from brain images. Traditional CNN architectures, like LeNet-5 and AlexNet, excel in capturing intricate structural information but may struggle with deeper, more abstract features. In contrast, transfer learning, where pre-trained models are fine-tuned for AD detection, has proven highly effective. This approach capitalizes on the wealth of knowledge acquired from diverse image datasets, offering a robust foundation for AD-related feature extraction. The introduction of 3D CNNs has further illuminated the importance of spatial context in AD diagnosis. These models excel in capturing nuanced patterns across multiple image slices, aligning with the progressive nature of AD. Performance metrics, including accuracy, sensitivity, specificity, and precision, have substantiated CNN’s effectiveness. These metrics provide quantitative evidence of CNNs’ diagnostic capabilities. Additionally, ongoing efforts in developing attention mechanisms and visualization tools aim to enhance model interpretability. However, the ultimate insight gleaned from CNN-based AD detection is the need for substantial data. Generalizability across diverse populations demands large, representative datasets. This challenge underscores the importance of data acquisition and curation efforts. In conclusion, CNNs have illuminated the path towards more accurate, data-driven AD detection. Leveraging hierarchical feature extraction, embracing 3D architectures, and ensuring interpretability are pivotal in harnessing CNNs’ potential for earlier and more reliable AD diagnosis.3.2. Recurrent Neural Networks (RNN) for AD Detection
Recurrent neural networks (RNNs) have acquired considerable attention in medical imaging for AD detection. These deep learning models are well-suited for capturing temporal dependencies and sequential patterns in data, making them particularly useful for analyzing time series or sequential data in AD detection tasks. In recent years, RNNs have shown promising results in capturing complex relationships within longitudinal neuroimaging data and aiding in the early diagnosis of AD.3.2.1. Long Short-Term Memory (LSTM) Networks
LSTM is a type of RNN that can effectively capture long-term dependencies in sequential data. Several studies have employed LSTM networks for AD diagnosis and prediction. These models typically take sequential data, such as time-series measurements from brain imaging or cognitive assessments, as input, and learn temporal patterns to classify or predict AD progression. LSTM-based models have demonstrated promising results in accurately diagnosing AD and predicting cognitive decline.3.2.2. Encoder–Decoder Architectures
Encoder–decoder architectures, often combined with attention mechanisms, have been used in AD research to address tasks such as predicting disease progression or generating informative features. These models encode input sequences into latent representations and decode them to generate predictions or reconstructed sequences. Encoder–decoder architectures with attention mechanisms allow for the network to focus on relevant temporal information, improving prediction accuracy and interpretability.3.2.3. Hybrid Models
Some studies have combined RNNs with other deep learning architectures, such as convolutional neural networks (CNNs) or generative adversarial networks (GANs), to leverage their respective strengths. These hybrid models aim to capture both spatial and temporal information from brain imaging data, leading to improved performance in AD diagnosis, progression prediction, or generating synthetic data for augmentation.3.2.4. Recurrent Neural Network (RNN) Studies for AD Detection
Recurrent neural networks (RNNs) have emerged as a popular deep learning technique for analyzing temporal data, making them well-suited for Alzheimer’s disease research. This discussion section will highlight the various methods that have utilized RNNs in AD research, provide an overview of their approaches, compare their performance, and present meaningful insights for further discussion.3.2.5. Performance Comparison
Comparing the performance of different RNN-based methods in AD research can be challenging due to variations in datasets, evaluation metrics, and experimental setups. However, several studies have reported high accuracy, sensitivity, and specificity in AD diagnosis and prediction tasks using RNNs. For example, LSTM-based models have achieved accuracies ranging from 80% to over 90% in AD classification. TCNs have demonstrated competitive performance in predicting cognitive decline, with high AUC scores. Encoder–decoder architectures with attention mechanisms have shown improvements in disease progression prediction compared to traditional LSTM models. Hybrid models combining RNNs with other architectures have reported enhanced performance by leveraging spatial and temporal information.3.2.6. Meaningful Insights
RNNs, such as LSTMs, are well-suited for capturing long-term dependencies in sequential data. In the context of AD research, this capability allows for identifying subtle temporal patterns and predicting disease progression. By analyzing longitudinal data, RNNs can potentially detect early signs of cognitive decline and facilitate early intervention strategies. RNNs can also be effectively used for data augmentation in AD research. Synthetic sequences can be generated using generative models, such as variational autoencoders (VAEs) or GANs, to increase the diversity and size of the training dataset. This augmented data can enhance the robustness and generalizability of RNN models, leading to improved diagnostic accuracy and generalization to unseen data. In addition, RNNs offer interpretability and explainability in AD research. By analyzing the temporal patterns learned by the models, researchers can gain insights into the underlying disease progression mechanisms. This information can aid in understanding the neurobiological processes associated with AD and provide valuable clues for potential therapeutic interventions. Moreover, RNNs can handle multimodal data sources, such as combining brain imaging (e.g., MRI, PET scans) with clinical assessments or genetic information. Integrating multiple modalities can provide a more comprehensive understanding of AD, capturing both structural and functional changes in the brain along with clinical markers. RNN-based models enable the fusion of diverse data sources to improve the accuracy and reliability of AD diagnosis and prognosis. RNNs trained on large-scale datasets can learn robust representations that generalize well to unseen data. Pre-training RNN models on large cohorts or external datasets and finetuning them on specific AD datasets can facilitate knowledge transfer and enhance the performance of AD classification and prediction tasks. Transfer learning approaches enable the utilization of existing knowledge and leverage the expertise gained from related tasks or domains. While RNNs have shown promise in AD research, there are still challenges to address. One major challenge is the limited availability of large-scale, longitudinal AD datasets. Acquiring and curating diverse datasets with longitudinal follow-up is crucial for training RNN models effectively. Additionally, incorporating uncertainty estimation and quantifying model confidence in predictions can further enhance the reliability and clinical applicability of RNN-based methods. Furthermore, exploring the combination of RNNs with other advanced techniques, such as attention mechanisms, graph neural networks, or reinforcement learning, holds promise for improving AD diagnosis, understanding disease progression, and guiding personalized treatment strategies. Integrating multimodal data sources, such as imaging, genetics, and omics data, can provide a more comprehensive view of AD pathophysiology.3.3. Generative Modeling for AD Detection
Generative modelling techniques have gained attention in medical imaging for Alzheimer’s disease (AD) detection. These models are capable of generating new samples that follow the distribution of the training data, enabling them to capture the underlying patterns and variations in AD-related imaging data. By leveraging generative models, researchers aim to enhance early detection, improve classification accuracy, and gain insights into the underlying mechanisms of AD.
3.3.1. GANs for Image Generation
One prominent application of generative modelling in Alzheimer’s disease is the generation of synthetic brain images for diagnostic and research purposes. GANs have been used to generate realistic brain images that mimic the characteristics of Alzheimer’s disease, such as the presence of amyloid beta plaques and neurofibrillary tangles. These synthetic images can be valuable for augmenting datasets, addressing data scarcity issues, and improving classification performance.5.3.2. Conditional GANs for Disease Progression Modeling
Conditional GANs (cGANs) have been employed to model the progression of Alzheimer’s disease over time. By conditioning the generator on longitudinal data, cGANs can generate synthetic brain images that capture disease progression stages, ranging from normal to mild cognitive impairment (MCI) and finally to Alzheimer’s disease. This enables the generation of realistic images representing the transition from healthy to pathological brain states.3.3.3. Variational Autoencoders (VAEs) for Feature Extraction
In addition to GANs, variational autoencoders (VAEs) have been utilized to extract informative features from brain images for Alzheimer’s disease classification. VAEs can learn a compressed representation of the input images, known as latent space, which captures relevant features associated with the disease. By sampling from the latent space, new images can be generated, and the extracted features can be used for classification tasks.3.3.4. Hybrid Approaches
Some studies have explored hybrid approaches that combine different generative models to leverage their respective advantages. For example, combining GANs and VAEs can harness the generative power of GANs while benefiting from the probabilistic nature and interpretability of VAEs. These hybrid models aim to generate high-quality images while preserving the meaningful representations learned by VAEs.3.3.5. Generative Modeling Studies (GAN) for AD Detection
Generative modelling, particularly through approaches like generative adversarial networks (GANs), has emerged as a promising technique in the field of Alzheimer’s disease research. This discussion section will provide an overview of the various methods used in generative modelling for Alzheimer’s disease, compare their strengths and limitations, and highlight meaningful insights for further exploration and discussion.3.3.6. Comparative Analysis
When comparing the different generative modelling methods in Alzheimer’s disease research, several factors should be considered:-
Image Quality: The primary goal of generative modelling is to generate high-quality brain images that closely resemble real data. GANs have demonstrated remarkable success in producing visually realistic images, while VAEs tend to produce slightly blurred images due to the nature of their probabilistic decoding process.
-
Feature Extraction: While GANs excel in image generation, VAEs are more suitable for feature extraction and latent space representation. VAEs can capture meaningful features that reflect disease progression and provide interpretability, making them valuable for understanding the underlying mechanisms of Alzheimer’s disease.
-
Data Scarcity: Alzheimer’s disease datasets are often limited in size, posing challenges for training deep learning models. Generative modelling techniques, especially GANs, can help address data scarcity by generating synthetic samples that augment the training data and improve model generalization.
-
Interpretability: VAEs offer an advantage in terms of interpretability because they learn a structured latent space that captures meaningful variations in the data. This can aid in understanding disease patterns and identifying potential biomarkers.
3.3.7. Meaningful Insights
Generative modelling in Alzheimer’s disease research holds great promise for advancing diagnosis, disease progression modelling, and understanding the underlying mechanisms of the disease. By generating realistic brain images and capturing disease-related features, these techniques can complement traditional diagnostic methods and provide new avenues for personalized treatment and intervention strategies. One meaningful insight from the application of generative modelling is the potential to address data scarcity issues. Alzheimer’s disease datasets are often limited in size and subject to variability in imaging protocols and data acquisition. By using generative models like GANs and VAEs, researchers can generate synthetic data that closely resemble real brain images. This augmentation of the dataset not only increases the sample size but also captures a wider range of disease characteristics and progression patterns. Consequently, it enhances the robustness and generalizability of machine learning models trained on these augmented datasets. Moreover, generative modelling techniques provide a unique opportunity to simulate disease progression and explore hypothetical scenarios. By conditioning the generative models on various disease stages, researchers can generate synthetic brain images that represent different pathological states, from early stages of mild cognitive impairment to advanced Alzheimer’s disease. This capability allows for the investigation of disease progression dynamics, identification of critical biomarkers, and evaluation of potential intervention strategies. Furthermore, the combination of generative models with other deep learning techniques, such as convolutional neural networks (CNNs) or recurrent neural networks (RNNs), can further enhance the performance of Alzheimer’s disease classification and prediction tasks. These hybrid models can leverage the strengths of different architectures and generate more accurate and interpretable results. For example, combining GANs for image generation with CNNs for feature extraction and classification can lead to improved diagnostic accuracy and a better understanding of the underlying disease mechanisms.References
- Vaupel, J.W. Biodemography of human ageing. Nature 2010, 464, 536–542.
- Samir, K.C.; Lutz, W. The human core of the shared socioeconomic pathways: Population scenarios by age, sex and level of education for all countries to 2100. Glob. Environ. Chang. 2017, 42, 181–192.
- Godyń, J.; Jończyk, J.; Panek, D.; Malawska, B. Therapeutic strategies for Alzheimer’s disease in clinical trials. Pharmacol. Rep. 2016, 68, 127–138.
- Blaikie, L.; Kay, G.; Lin, P.K.T. Current and emerging therapeutic targets of Alzheimer’s disease for the design of multi-target directed ligands. MedChemComm 2019, 10, 2052–2072.
- Gutierrez, B.A.; Limon, A. Synaptic disruption by soluble oligomers in patients with Alzheimer’s and Parkinson’s disease. Biomedicines 2022, 10, 1743.
- Zvěřová, M. Clinical aspects of Alzheimer’s disease. Clin. Biochem. 2019, 72, 3–6.
- Bronzuoli, M.R.; Iacomino, A.; Steardo, L.; Scuderi, C. Targeting neuroinflammation in Alzheimer’s disease. J. Inflamm. Res. 2016, 9, 199–208.
- Aljunid, S.M.; Maimaiti, N.; Ahmed, Z.; Nur, A.M.; Nor, N.M.; Ismail, N.; Haron, S.A.; Shafie, A.A.; Salleh, M.; Yusuf, S.; et al. Development of clinical pathway for mild cognitive impairment and dementia to quantify cost of age-related cognitive disorders in Malaysia. Malays. J. Public Health Med. 2014, 14, 88–96.
- Hu, K.; Wang, Y.; Chen, K.; Hou, L.; Zhang, X. Multi-scale features extraction from baseline structure MRI for MCI patient classification and AD early diagnosis. Neurocomputing 2016, 175, 132–145.
- Ebrahimighahnavieh, A.; Luo, S.; Chiong, R. Deep learning to detect Alzheimer’s disease from neuroimaging: A systematic literature review. Comput. Methods Programs Biomed. 2020, 187, 105242.
- Fathi, S.; Ahmadi, M.; Dehnad, A. Early diagnosis of Alzheimer’s disease based on deep learning: A systematic review. Comput. Biol. Med. 2022, 146, 105634.
- Binaco, R.; Calzaretto, N.; Epifano, J.; McGuire, S.; Umer, M.; Emrani, S.; Wasserman, V.; Libon, D.J.; Polikar, R. Machine learning analysis of digital clock drawing test performance for differential classification of mild cognitive impairment subtypes versus Alzheimer’s disease. J. Int. Neuropsychol. Soc. 2020, 26, 690–700.
- Karikari, T.K.; Ashton, N.J.; Brinkmalm, G.; Brum, W.S.; Benedet, A.L.; Montoliu-Gaya, L.; Lantero-Rodriguez, J.; Pascoal, T.A.; Suárez-Calvet, M.; Rosa-Neto, P.; et al. Blood phospho-tau in Alzheimer disease: Analysis, interpretation, and clinical utility. Nat. Rev. Neurol. 2022, 18, 400–418.
- Wang, M.; Song, W.M.; Ming, C.; Wang, Q.; Zhou, X.; Xu, P.; Krek, A.; Yoon, Y.; Ho, L.; Orr, M.E.; et al. Guidelines for bioinformatics of single-cell sequencing data analysis in Alzheimer’s disease: Review, recommendation, implementation and application. Mol. Neurodegener. 2022, 17, 1–52.
- Zhang, Y.; Wu, K.-M.; Yang, L.; Dong, Q.; Yu, J.-T. Tauopathies: New perspectives and challenges. Mol. Neurodegener. 2022, 17, 28.
- Klyucherev, T.O.; Olszewski, P.; Shalimova, A.A.; Chubarev, V.N.; Tarasov, V.V.; Attwood, M.M.; Syvänen, S.; Schiöth, H.B. Advances in the development of new biomarkers for Alzheimer’s disease. Transl. Neurodegener. 2022, 11, 1–24.
- Zhao, K.; Duka, B.; Xie, H.; Oathes, D.J.; Calhoun, V.; Zhang, Y. A dynamic graph convolutional neural network framework reveals new insights into connectome dysfunctions in ADHD. NeuroImage 2022, 246, 118774.
- Jiang, Y.; Zhou, X.; Ip, F.C.; Chan, P.; Chen, Y.; Lai, N.C.; Cheung, K.; Lo, R.M.; Tong, E.P.; Wong, B.W.; et al. Large-scale plasma proteomic profiling identifies a high-performance biomarker panel for Alzheimer’s disease screening and staging. Alzheimer’s Dement. 2022, 18, 88–102.
- Gómez-Isla, T.; Frosch, M.P. Lesions without symptoms: Understanding resilience to Alzheimer disease neuropathological changes. Nat. Rev. Neurol. 2022, 18, 323–332.
- Liu, C.; Xiang, X.; Han, S.; Lim, H.Y.; Li, L.; Zhang, X.; Ma, Z.; Yang, L.; Guo, S.; Soo, R.; et al. Blood-based liquid biopsy: Insights into early detection and clinical management of lung cancer. Cancer Lett. 2022, 524, 91–102.
- Hernandez, M.; Ramon-Julvez, U.; Ferraz, F.; with the ADNI Consortium. Explainable AI toward understanding the performance of the top three TADPOLE Challenge methods in the forecast of Alzheimer’s disease diagnosis. PLoS ONE 2022, 17, e0264695.
- Lydon, E.A.; Nguyen, L.T.; Shende, S.A.; Chiang, H.-S.; Spence, J.S.; Mudar, R.A. EEG theta and alpha oscillations in early versus late mild cognitive impairment during a semantic Go/NoGo task. Behav. Brain Res. 2022, 416, 113539.
- Yadav, S.; Zhou Shu, K.; Zachary, Z.; Yueyang, G.; Lana, X.; ADNI Consortium. Integrated Metabolomics and Transcriptomics Analysis Identifies Molecular Subtypes within the Early and Late Mild Cognitive Impairment Stages of Alzheimer’s Disease. medRxiv 2023.
- Jeyavathana, R.B.; Balasubramanian, R.; Pandian, A.A. A survey: Analysis on pre-processing and segmentation techniques for medical images. Int. J. Res. Sci. Innov. (IJRSI) 2016, 3, 113–120.
- James, A.P.; Dasarathy, B.V. Medical image fusion: A survey of the state of the art. Inf. Fusion 2014, 19, 4–19.
- Balagurunathan, Y.; Mitchell, R.; El Naqa, I. Requirements and reliability of AI in the medical context. Phys. Medica 2021, 83, 72–78.
- Waldemar, G.; Dubois, B.; Emre, M.; Georges, J.; McKeith, I.G.; Rossor, M.; Scheltens, P.; Tariska, P.; Winblad, B. Recommendations for the diagnosis and management of Alzheimer’s disease and other disorders associated with dementia: EFNS guideline. Eur. J. Neurol. 2007, 14, e1–e26.
- Shenton, M.E.; Hamoda, H.M.; Schneiderman, J.S.; Bouix, S.; Pasternak, O.; Rathi, Y.; Vu, M.-A.; Purohit, M.P.; Helmer, K.; Koerte, I.; et al. A review of magnetic resonance imaging and diffusion tensor imaging findings in mild traumatic brain injury. Brain Imaging Behav. 2012, 6, 137–192.
- Matthews, P.M.; Peter, J. Functional magnetic resonance imaging. J. Neurol. Neurosurg. Psychiatry 2004, 75, 6–12.
- Klunk, W.E.; Mathis, C.A.; Price, J.C.; DeKosky, S.T.; Lopresti, B.J.; Tsopelas, N.D.; Judith, A.S.; Robert, D.N. Amyloid imaging with PET in Alzheimer’s disease, mild cognitive impairment, and clinically unimpaired subjects. In PET in the Evaluation of Alzheimer’s Disease and Related Disorders; Springer: New York, NY, USA, 2009; pp. 119–147.
- Basser, P.J. Inferring microstructural features and the physiological state of tissues from diffusion-weighted images. NMR Biomed. 1995, 8, 333–344.
- Braak, H.; Alafuzoff, I.; Arzberger, T.; Kretzschmar, H.; Del Tredici, K. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 2006, 112, 389–404.
- Dubois, B.; Feldman, H.H.; Jacova, C.; DeKosky, S.T.; Barberger-Gateau, P.; Cummings, J.L.; Delacourte, A.; Galasko, D.; Gauthier, S.; Jicha, G.A.; et al. Research criteria for the diagnosis of Alzheimer’s disease: Revising the NINCDS–ADRDA criteria. Lancet Neurol. 2007, 6, 734–746.
- Mueller, S.G.; Weiner, M.W.; Thal, L.J.; Petersen, R.C.; Jack, C.; Jagust, W.; Trojanowski, J.Q.; Toga, A.W.; Beckett, L. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin. 2005, 15, 869–877.
- Jagust, W. Imaging the evolution and pathophysiology of Alzheimer disease. Nat. Rev. Neurosci. 2018, 19, 687–700.
- Weiner, M.W.; Veitch, D.P.; Aisen, P.S.; Beckett, L.A.; Cairns, N.J.; Green, R.C.; Harvey, D.; Jack, C.R.; Jagust, W.; Liu, E.; et al. The Alzheimer’s Disease Neuroimaging Initiative: A review of papers published since its inception. Alzheimer’s Dement. 2013, 9, e111–e194.
- Klein, A.; Andersson, J.; Ardekani, B.A.; Ashburner, J.; Avants, B.; Chiang, M.-C.; Christensen, G.E.; Collins, D.L.; Gee, J.; Hellier, P.; et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. NeuroImage 2009, 46, 786–802.
- Ashburner, J.; Friston, K.J. Voxel-based morphometry—The methods. Neuroimage 2000, 11, 805–821.
- Choy, G.; Khalilzadeh, O.; Michalski, M.; Synho, D.; Samir, A.E.; Pianykh, O.S.; Geis, J.R.; Pandharipande, P.V.; Brink, J.A.; Dreyer, K.J. Current applications and future impact of machine learning in radiology. Radiology 2018, 288, 318–328.
- Smith, S.M.; Jenkinson, M.; Woolrich, M.W.; Beckmann, C.F.; Behrens, T.E.; Johansen-Berg, H.; Bannister, P.R.; De Luca, M.; Drobnjak, I.; Flitney, D.E.; et al. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage 2004, 23, S208–S219.
- Jenkinson, M.; Bannister, P.; Brady, M.; Smith, S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 2002, 17, 825–841.
- Hashem IA, T.; Yaqoob, I.; Anuar, N.B.; Mokhtar, S.; Gani, A.; Khan, S.U. The rise of “big data” on cloud computing: Review and open research issues. Inf. Syst. 2015, 47, 98–115.
- Gorgolewski, K.J.; Auer, T.; Calhoun, V.D.; Craddock, R.C.; Das, S.; Duff, E.P.; Flandin, G.; Ghosh, S.S.; Glatard, T.; Halchenko, Y.O.; et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data 2016, 3, 160044.
- Kandel, S.; Heer, J.; Plaisant, C.; Kennedy, J.; van Ham, F.; Riche, N.H.; Weaver, C.; Lee, B.; Brodbeck, D.; Buono, P. Research directions in data wrangling: Visualizations and transformations for usable and credible data. Inf. Vis. 2011, 10, 271–288.
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444.
- Madrid, L.; Labrador, S.C.; González-Pérez, A.; Sáez, M.E.; Alzheimer’s Disease Neuroimaging Initiative (ADNI and others). Integrated Genomic, Transcriptomic and Proteomic Analysis for Identifying Markers of Alzheimer’s Disease. Diagnostics 2021, 11, 2303.
- Konur, O.; Kingma, D.; Ba, J. Adam: A method for stochastic optimization. In Proceedings of the International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, 7–9 May 2015; pp. 1–15.
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958.
- Jiang, X.; Chang, L.; Zhang, Y.-D. Classification of Alzheimer’s disease via eight-layer convolutional neural network with batch normalization and dropout techniques. J. Med. Imaging Health Inform. 2020, 10, 1040–1048.
- Li, Z.; Liu, F.; Yang, W.; Peng, S.; Zhou, J. A survey of convolutional neural networks: Analysis, applications, and prospects. IEEE Trans. Neural Netw. Learn. Syst. 2021, 33, 6999–7019.
- Bin Bae, J.; Lee, S.; Jung, W.; Park, S.; Kim, W.; Oh, H.; Han, J.W.; Kim, G.E.; Kim, J.S.; Kim, J.H.; et al. Identification of Alzheimer’s disease using a convolutional neural network model based on T1-weighted magnetic resonance imaging. Sci. Rep. 2020, 10, 22252.
- Ilesanmi, A.E.; Ilesanmi, T.; Idowu, O.P.; Torigian, D.A.; Udupa, J.K. Organ segmentation from computed tomography images using the 3D convolutional neural network: A systematic review. Int. J. Multimed. Inf. Retr. 2022, 11, 315–331.
- Irshad, S.; Gomes, D.P.S.; Kim, S.T. Improved abdominal multi-organ segmentation via 3d boundary-constrained deep neural networks. IEEE Access 2023, 11, 35097–35110.
- Rickmann, A.-M.; Senapati, J.; Kovalenko, O.; Peters, A.; Bamberg, F.; Wachinger, C. AbdomenNet: Deep neural network for abdominal organ segmentation in epidemiologic imaging studies. BMC Med. Imaging 2022, 22, 1–11.
- Hassan, S.M.; Maji, A.K. Plant disease identification using a novel convolutional neural network. IEEE Access 2022, 10, 5390–5401.
- Ashwinkumar, S.; Rajagopal, S.; Manimaran, V.; Jegajothi, B. Automated plant leaf disease detection and classification using optimal MobileNet based convolutional neural networks. Mater. Today Proc. 2022, 51, 480–487.
- Jiang, J.; Liu, H.; Zhao, C.; He, C.; Ma, J.; Cheng, T.; Zhu, Y.; Cao, W.; Yao, X. Evaluation of Diverse Convolutional Neural Networks and Training Strategies for Wheat Leaf Disease Identification with Field-Acquired Photographs. Remote Sens. 2022, 14, 3446.
- Nirthika, R.; Manivannan, S.; Ramanan, A.; Wang, R. Pooling in convolutional neural networks for medical image analysis: A survey and an empirical study. Neural Comput. Appl. 2022, 34, 5321–5347.
- Mattia, G.M.; Sarton, B.; Villain, E.; Vinour, H.; Ferre, F.; Buffieres, W.; Le Lann, M.-V.; Franceries, X.; Peran, P.; Silva, S. Multimodal MRI-Based Whole-Brain Assessment in Patients In Anoxoischemic Coma by Using 3D Convolutional Neural Networks. Neurocritical Care 2022, 37 (Suppl. S2), 303–312.
- Warren, S.L.; Moustafa, A.A. Functional magnetic resonance imaging, deep learning, and Alzheimer’s disease: A systematic review. J. Neuroimaging 2023, 33, 5–18.
- Dan, T.; Huang, Z.; Cai, H.; Laurienti, P.J.; Wu, G. Learning brain dynamics of evolving manifold functional MRI data using geometric-attention neural network. IEEE Trans. Med. Imaging 2022, 41, 2752–2763.
- Guo, X.; Zhou, B.; Pigg, D.; Spottiswoode, B.; Casey, M.E.; Liu, C.; Dvornek, N.C. Unsupervised inter-frame motion correction for whole-body dynamic PET using convolutional long short-term memory in a convolutional neural network. Med. Image Anal. 2022, 80, 102524.
- Bin Tufail, A.; Anwar, N.; Ben Othman, M.T.; Ullah, I.; Khan, R.A.; Ma, Y.-K.; Adhikari, D.; Rehman, A.U.; Shafiq, M.; Hamam, H. Early-stage Alzheimer’s disease categorization using PET neuroimaging modality and convolutional neural networks in the 2d and 3d domains. Sensors 2022, 22, 4609.
- Estudillo-Romero, A.; Haegelen, C.; Jannin, P.; Baxter, J.S.H. Voxel-based diktiometry: Combining convolutional neural networks with voxel-based analysis and its application in diffusion tensor imaging for Parkinson’s disease. Hum. Brain Mapp. 2022, 43, 4835–4851.
- Liu, S.; Liu, Y.; Xu, X.; Chen, R.; Liang, D.; Jin, Q.; Liu, H.; Chen, G.; Zhu, Y. Accelerated cardiac diffusion tensor imaging using deep neural network. Phys. Med. Biol. 2022, 68, 025008.
- Park, S.; Yu, J.; Woo, H.-H.; Park, C.G. A novel network architecture combining central-peripheral deviation with image-based convolutional neural networks for diffusion tensor imaging studies. J. Appl. Stat. 2022, 50, 3294–3311.
- AlSaeed, D.; Omar, S.F. Brain MRI analysis for Alzheimer’s disease diagnosis using CNN-based feature extraction and machine learning. Sensors 2022, 22, 2911.
- Ghazal, T.M.; Abbas, S.; Munir, S.; Ahmad, M.; Issa, G.F.; Zahra, S.B.; Khan, M.A.; Hasan, M.K. Alzheimer disease detection empowered with transfer learning. Comput. Mater. Contin. 2022, 70, 5005–5019.
- Liu, S.; Masurkar, A.V.; Rusinek, H.; Chen, J.; Zhang, B.; Zhu, W.; Fernandez-Granda, C.; Razavian, N. Generalizable deep learning model for early Alzheimer’s disease detection from structural MRIs. Sci. Rep. 2022, 12, 17106.
- Loddo, A.; Buttau, S.; Di Ruberto, C. Deep learning based pipelines for Alzheimer’s disease diagnosis: A comparative study and a novel deep-ensemble method. Comput. Biol. Med. 2021, 141, 105032.
 Encyclopedia
Encyclopedia
