Brain tumor segmentation plays a crucial role in the diagnosis, treatment planning, and monitoring of brain tumors. Accurate segmentation of brain tumor regions from multi-sequence magnetic resonance imaging (MRI) data is of paramount importance for precise tumor analysis and subsequent clinical decision making. The ability to delineate tumor boundaries in MRI scans enables radiologists and clinicians to assess tumor size, location, and heterogeneity, facilitating treatment planning and evaluating treatment response. Traditional manual segmentation methods are time-consuming, subjective, and prone to inter-observer variability. Therefore, the automatic segmentation algorithm has received widespread attention as an alternative solution. For instance, the self-organizing map (SOM) is an unsupervised exploratory data analysis tool that leverages principles of vector quantization and similarity measurement to automatically partition images into self-similar regions or clusters. Segmentation methods based on SOM have demonstrated the ability to distinguish high-level and low-level features of tumors, edema, necrosis, cerebrospinal fluid, and healthy tissue.
1. Attention Mechanisms in Convolutional Neural NetworkNs
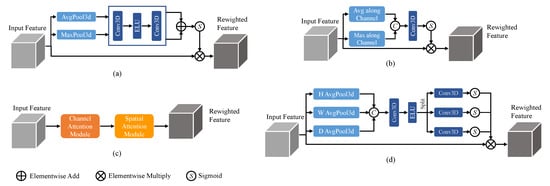
The channel attention mechanism
[1][21] focuses on enhancing important channel information while suppressing less relevant channels, achieved by computing attention weights along the channel dimension. It typically involves global average pooling and a multi-layer perceptron (MLP) to generate attention weights, which are then used to weight the original feature maps. On the other hand, the spatial attention mechanism
[2][20] aims to highlight important spatial locations and suppress unimportant ones. It can be implemented through convolutional operations with different kernel sizes or using self-attention mechanisms to compute attention weights between spatial positions. These attention mechanisms, when combined, form the Convolutional Block Attention Module (CBAM)
[3][36], which integrates both channel and spatial attention. Additionally, a recently proposed attention mechanism called coordinate attention
[4][37] focuses on modeling the relationships between different positions in the feature maps by incorporating coordinate information. It utilizes an MLP to process the coordinate information and generates position weights, which are then multiplied with the original feature maps. The 3D structures of these commonly used attention mechanisms are illustrated in
Figure 1. While these attention mechanisms enhance the modeling capability of CNNs by capturing important channel, spatial, and positional information, they also introduce additional computational overhead.
Figure 1.
Attention mechanisms with (
a
) channel attention, (
b
) spatial attention, (
c
) CBAM, and (
d
) coordinate attention.
2. Single-Path and Multi-Path Convolutional Neural NetworkNNs for Brain Tumor Segmentation
CNNs have shown great promise in achieving accurate and efficient segmentation results, revolutionizing the way brain tumors are analyzed and diagnosed due to their ability to automatically learn discriminating features from input data
[5][38]. Initially, single-path CNNs were employed, where a single data processing stream was utilized
[6][39]. These networks, which take multi-modal brain tumor
magnetic resonance imaging (MRI
) scans as input, sequentially pass the data through a combination of convolutional layers, pooling layers, and non-linear activation layers, ultimately performing segmentation using a classifier at the end of the model. Single-path CNNs are characterized by their simplicity in structure and shallow hierarchy, but their segmentation performance may be sub-optimal. As brain tumor images are inherently complex and diverse, relying solely on a single processing path may limit the network’s ability to capture and represent the intricate details present in different modalities.
To address this limitation, multi-path CNNs have been introduced, featuring multiple parallel convolutional pathways
[7][40]. This architecture allows for the processing of input information at multiple scales, providing a larger receptive field and the potential for enhanced segmentation accuracy. However, it is worth noting that multi-path CNNs tend to exhibit a higher level of complexity and require a larger model size to accommodate the increased number of pathways. Furthermore, an inherent challenge arises from the uneven distribution of tumor regions, where certain tumor areas may exhibit varying sizes and proportions compared to others.
To tackle this class imbalance issue, cascaded CNNs have been proposed as a potential solution
[8][41]. By cascading multiple network models, each designed to segment a specific region of interest, cascaded CNNs enable the transformation of the multi-region tumor segmentation problem into a series of binary segmentation tasks. One of the key advantages of cascaded CNNs is their ability to consider the unique relationships between sub-regions when predicting subsequent segmentation tasks. This can be particularly beneficial in minimizing false positives, as each network operates on regions extracted from the output of the previous network. However, an important point to consider is that cascaded CNNs, in contrast to single-path and multi-path CNNs, are not end-to-end and require additional time for training and testing due to the sequential nature of the cascaded segmentation process.
3. The U-Net and Its Variants for Brain Tumor Segmentation
The U-Net architecture consists of an encoder–decoder structure coupled with skip connections
[9][10]. The encoder path incorporates a series of convolutional and pooling layers to progressively extract hierarchical features and reduce spatial resolution. The decoder path utilizes up-sampling and transposed convolutional layers to recover the spatial information and generate segmentation maps. Skip connections connect the corresponding encoder and decoder layers, allowing for the model to fuse low-level and high-level features. This design enables U-Net to capture both local and global contextual information, facilitating the accurate delineation of tumor boundaries.
Initially, the research focused on 2D segmentation networks operating within individual 2D image planes. U-Net
[10][12] has demonstrated its efficacy in capturing tumor boundaries and distinguishing tumor regions from healthy brain tissue. U-Net++
[11][25] extends the U-Net architecture by incorporating nested and dense skip pathways, enabling the capture of multi-scale contextual information for precise brain tumor segmentation. SegResNet
[12][26] combines U-Net architecture with the residual network (ResNet) to enhance feature representation and segmentation performance, effectively capturing both local and global contextual information. To further improve feature representation, DynU-Net
[13][27] integrates a dynamic routing algorithm inspired by capsule networks into the U-Net architecture, enabling the capture of hierarchical relationships among different tumor regions. MS-Net
[14][31] is a medical image segmentation technique based on a codec structure composed of a Multi-Scale Attention Module (MSAM) and a Stacked Feature Pyramid Module (SFPM). MSAM dynamically adjusts the receptive fields to capture different levels of context details, while SFPM adaptively increases the weight of the features of interest to focus the network’s attention on the target region. Fusion factor
[15][42] is introduced to control the amount of information transferred from deep to shallow layers in Feature Pyramid Networks (FPN) for tiny object detection. The paper explores how to estimate the effective value of the fusion factor for a specific dataset by statistical methods. However, these 2D networks may disregard the crucial depth information inherent in the MRI images, consequently impeding their ability to comprehensively utilize the rich local and global contextual information available.
Therefore, 3D U-Net
[16][43] was developed to extend the U-Net framework for processing volumetric data, enabling the segmentation of brain tumors in 3D medical images. By considering spatial dependencies along the three dimensions, SCAR U-Net
[17][44] improves the accuracy of tumor segmentation in volumetric scans. V-Net
[18][45] is another extension of U-Net that incorporates a volumetric residual learning framework. It leverages 3D convolutional neural networks and residual connections to capture fine-grained details in volumetric data. The evolution of mainstream 2D segmentation networks into their 3D counterparts has resulted in significant improvements in brain tumor segmentation performance
[19][46]. DSTGAN
[20][47] presents a spatiotemporal generative adversarial learning approach for segmentation and quantification of myocardial infarction without contrast agents. The approach utilizes a generator and a discriminator module, which consist of three seamlessly connected networks to extract the morphological and motion abnormalities of the left ventricle, learn the complementarity between segmentation and quantification tasks, and leverage adversarial learning to enhance the accuracy of estimation. However, it remains crucial to strike a balance between model complexity and computational feasibility, considering the practical constraints and available computational resources.
Simultaneously, the transformer
[21][48] architecture has gained significant popularity in natural language processing (NLP) and has found applications in medical image analysis
[22][23][49,50]. Initially developed for sequence modeling tasks, transformers have showcased their ability to capture long-range dependencies and capture contextual information effectively. Building upon this success, researchers have extended transformers to medical image analysis, leading to the emergence of models. UNETR
[24][30] combines the transformer architecture with the U-Net framework, enabling the modeling of long-range dependencies and achieving state-of-the-art performance in brain tumor segmentation. Similarly, SwinUNETR
[25][28] integrates the Swin Transformer, a hierarchical vision transformer, with the U-Net framework, effectively capturing global and local dependencies with reduced computational complexity. nnFormer
[26][32] is a novel approach using a 3D transformer to segment medical images based on interleaved convolution and self-attention operations. It introduces local and global volume-based self-attention to learn volume representations and outperforms previous transformer-based methods on three public datasets. SeMask
[27][51] proposes a semantically masked transformer network for semantic segmentation of images. The network leverages an additional semantic layer to incorporate semantic information about the image, which improves the performance of the pre-trained transformer backbone. However, these transformer-based U-Net models face challenges such as increased model size, longer training time, and higher computational requirements, which can limit their practicality in real-world applications.