DOG1 (Delay of Germination-1), is a master regulator of primary seed dormancy that acts in concert with ABA to delay germination. The strongly regulated expression of DOG1 increases during seed maturation, and the DOG1-mRNA disappears quickly after seed imbibition, although the DOG1 protein is more stable. DOG1 expression is notably induced by abscisic acid (ABA) and low temperature during seed maturation. The DOG1 action involves the suppression of the ABA HYPERSENSITIVE GERMINATION (AHG1/AHG3) activity to enhance ABA sensitivity and finally impose the primary dormancy (PD). This suppression needs the formation of DOG1-heme complex. Together, DOG1 function is not restricted to PD process, but that it is also required for other facets of seed maturation and plant development (e.g. flowering and drought tolerance), in part by also interfering with the ethylene signaling components.
- DOG1
- ABA
- Heme group
- miRNA
- AHG1
- Primary dormancy
1. Introduction
Throughout this study we have reviewed a good number of recent publications related to DOG1, a protein absolutely required for the induction of primary dormancy (PD). However, its molecular function is largely unknown. The hypothesis and conclusions that are currently underway in this review are largely based on data obtained from A. thaliana seed mutants and the characterization of cofactors that bind to different sites of the DOG1 structure. All known genetic and molecular data are part of a highly complex puzzle that, acting in a coordinated mode with abscisic acid (ABA), trigger the generation of active DOG1 and appearance of PD. However, although the function of DOG1 seems conserved in Angiosperms, much more experimentation in monocot and dicot species needs to be done to confirm. The key to addressing the emergence and evolution of DOG1 involves studying similar genes in the DOG1 family (Figure 1). Given the gaps existing in the knowledge of the mechanism of action of ABA in the PD process, this evolutionary study of DOG1 is of enormous importance since it firmly proves that DOG1 inhibits germination in an ABA-dependent way (Nishimura et al., 2018) [1]. However, ABA accumulates in seeds mainly to prevent viviparism and establish PD. The relationship between DOG1 and development of viviparism will be an important applied approach because reduced yield of certain crops is involved. On the other hand, the knowledge of endogenous and environmental signals responsible for the appearance/disappearance of DOG1 in the cells involved in the triggering, maintenance, and loss of PD, is an aspect still obscure and that requires a great deal of dedication. Recent research found that the production of miRNAs others than miR156 and miR172 (i.e., miR319, miR160, miR164, and miR390) are affected by DOG1 in lettuce and Arabidopsis (Huo et al., 2016) [2]. Although it seems clear that some miRNAs affect DOG1, it has not been conclusively proven yet that miRNA intervention is part of the primary mechanism by which DOG1 regulates PD. Hence, this intriguing and novel aspect should be taken into consideration to demonstrate the function of DOGs more comprehensively.
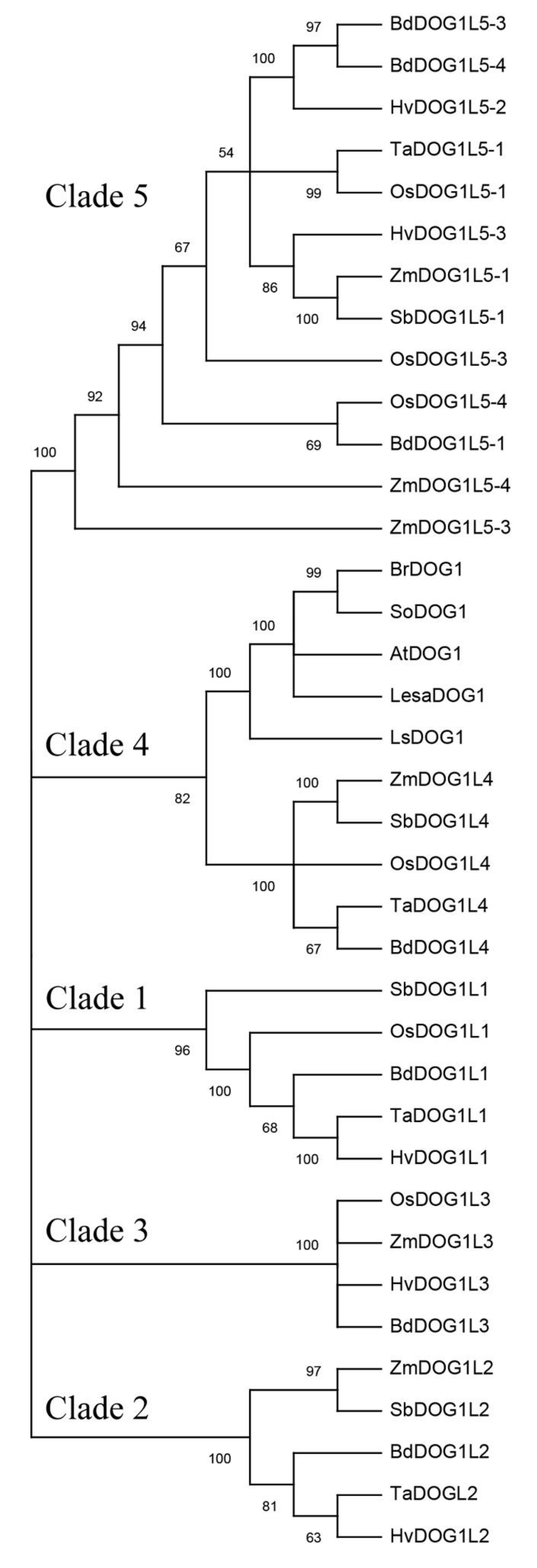
Figure 1. Phylogenetic tree of DOG1. Deduced DOG1 and DOG1-like amino acid sequences of Arabidopsis thaliana (AtDOG1, AtDOGL1-5); deduced DOG1 amino acid sequences of Brassica rapa (BrDOG1), Lactuca sativa (LsDOG1), Lepidum sativum (LesaDOG1), Sisymbrium officinale (SoDOG1); and the homologs in cereals Brachypodium distachyon (BdDOG1L1 to L5), Hordeum vulgare (HvDOG1L1, L2, L3, and L5), Oryza sativa (OsDOG1L1, L3, L4, and L5), Sorghum bicolor (SbDOG1L1 to L5), Triticum aestivum (TaDOG1L1, L2, L4, and L5), and Zea mays (ZmDOG1L2 to L5) were included. Tree was constructed with the neighbor joining method (1000 bootstrap repetitions) using MEGA X software. Only branches with a value above 50% are shown. An additional table indicates the accession numbers of the sequences included in the tree.
2. Perspectives for Future Research on DOG1
The yeast two-hybrid analysis (YTHA), co-immunoprecipitation, plant co-transformation, and epistatic analysis, are essential tools to progress in the knowledge of DOG1 at the molecular level. As a proof of the use of these methodologies, the breakthrough came from recent investigation about DOG1-interacting proteins (e.g., AHG1-DOG1). Although the interaction of DOG1 with ABA-Hipersensitive-Germination-1 (AHG1) seems demonstrated, it is still not clear if the inactivation of AHG1 by DOG1 in vivo is a consequence of that interaction. Given that DOG1 affects the expression level of ABI5, the DOG1-AHG1 complex likely regulates ABI5 function [3]. But this fact is still unsolved. Intriguingly, since the absorption spectrum of the active DOG1 exhibits characteristics of a heme-protein complex, it was proven that DOG1 binds to a heme group using His residues from its protein (Figure 2). It is striking that heme is not fundamental for DOG1 to interact with AHG1 (Nishimura et al., 2018; Nonogaki, 2020) [4]. Once it was known that heme group is involved in the action of DOG1, the PD mechanisms goes through a thorough investigation into the DOG1-heme relationship. Parallelly, at the evolutionary level, more research on ABA and heme signaling in the algal ancestor and bryophytes, are essential. However, the origin of heme in seeds is still an open question. Thus, is the plasmid only the source of heme in the seed cells or is there heme of mitochondrial origin. This question opens an interesting line of research since the heme group has a remarkable interference in many aspects of seed physiology. Further studies are needed to elucidate the potential role of heme bound to DOG1 as a sensor of O2 or NO. Thus, in the course of after-ripening (AR), several oxidative processes are produced, causing post-translational modifications in DOG1. It is necessary to investigate the possibility that ROS could oxidize DOG1. The nature of DOG1 modifications (e.g., phosphorylation/de-phosphorylation, redox changes, chromatin alterations, etc.,) in dry or imbibed AR viable seeds has not been demonstrated explicitly. These modifications may contribute to the change in the configuration of the DOG1 protein. Therefore, to carry out this approach, the study of the AR process in mutant seeds that are affected in the functioning of DOG1 can be an interesting tool (Chathane et al., 2017; Née et al., 2017)[5][6]. Finally, the knowledge in depth of PD physiology will imply to have tools for the control, among others, of crops with high agricultural value. In other words, the identification of DOG1 protein modifications is key to our understanding of PD and could enable the manipulation of PD levels in crop plants. Thus, a suitable understanding of PD will be advantageous for both ecological understanding and crop management. Likewise, it will be interesting to find out whether DOG1 is involved in the adaptation of PD to other environmental conditions that occur during seed maturation, like drought (Figure 3), light intensity, daylength, high or low temperatures, and nitrate levels. Together, the knowledge in depth of PD physiology will imply to have tools for the control, among others, of crops with high agricultural interest. Finally, to highlight the CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein) based genome editing approach has become a choice of technique due to its simplicity, ease of access, and flexibility [7]. The CRISPR/Cas9 system is a revolutionary technology for crop breeding and biological research through direct and controlled changes in the genome. Thus, the potential to edit multiple targets simultaneously makes CRIPSR/Cas9 possible to take up more challenging tasks required to engineer desired crop plants. The new gene editing techniques are more precise than standard genetic engineering tools that have been previously developed. The use of this technology will cooperate in the strong advance of knowledge of DOG1.
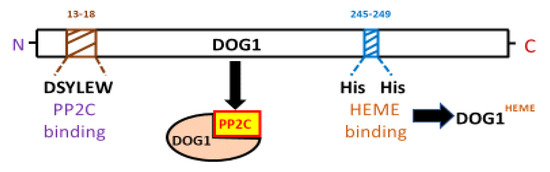
Figure 2. AtDOG1 protein structure indicating the positions of the AHG1 and heme binding sites. DOG1 is an α-helical protein that has the ability to bind both ABA HYPERSENSITIVE GERMINATION1 (AHG1; a clade A protein phosphatase 2C) and heme group. The amino acid residues that specifically bind PP2C (i.e., DSYLEW) are very close to the N-terminal; while those that specifically bind heme group are closer to the C-terminal. Heme binding is not necessary for DOG1 interaction with AHG1. However, heme binding at His245 and His249 is essential for DOG1 function in seed dormancy.
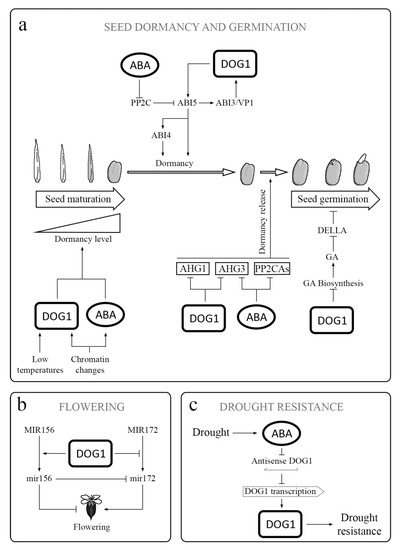
Figure 3. DOG1 in different physiological processes: (a) DOG1 has an influence on several genes involved in ABA signaling, including ABI3 and ABI5. Thus, it acts together with ABA in the regulation of the acquisition, maintenance, and release of PD. DOG1 also inactivates AHG1 and AHG3, which are the key negative regulators in ABA signaling. The inactivation of AHG1 and AHG2 results in release of PD. Additionally, DOG1 reduces the GAs biosynthesis during germination. (b) DOG1 participates in the regulation of flowering by influencing the transition from primary MIR156 and MIR172 to active miR156 and miR172 through an effect on expression of genes involved in miRNA processing. Both active miRNAs are regulators, not only of flowering but also of PD and seed and seedling development. (c) Under drought conditions, the ABA content increases. The expression of DOG1 enhances the drought tolerance in vegetative tissues, and asDOG1 transcription is ABA repressed. asDOG1 acts as a negative regulator of DOG1 transcription.
References
- Nishimura, N.; Tsuchiya, W.; Moresco, J.J.; Hayashi, Y.; Satoh, K.; Kaiwa, N.; Irisa, T.; Kinoshita, T.; Schroeder, J.I.; Yates, J.R.; et al. Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nat. Commun. 2018, 9, 2132.
- Huo, H.; Wei, S.; Bradford, K.J. DELAY OF GERMINATION1(DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc. Natl. Acad. Sci. USA 2016, 113, E2199–E2206.
- Dekkers, B.J.; He, H.; Hanson, J.; Willems, L.A.; Jamar, D.C.; Cueff, G.; Rajjou, L.; Hilhorst, H.W.; Bentsink, L. The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. Plant J. 2016, 85, 451–465
- Nonogaki, H. J. Exp. Bot. 2020, doi: 10.1093/jxb/eraao62.
- Chahtane, H.; Kim, W.; Lopez-Molina, L. Primary seed dormancy: A temporally multilayered riddle waiting to be unlocked. J. Exp. Bot. 2017, 68, 857–869.
- Née, G.; Kramer, K.; Nakabayashi, K.; Yuan, B.; Xiang, Y.; Miatton, E.; Finkemeier, I.; Soppe, W.J.J. DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signaling pathway to control seed dormancy. Nat. Commun. 2017, 8, 72.
- Corte, L.E.D.; Mhmaoud, L.M.; Moraes, T.S.; Mou, Z.; Grosser, J.W.; Dutt, M.; Development of Improved Fruit, Vegetable, and Ornamental Crops Using the CRISPR/Cas9 Genome Editing Technique. Lígia Erpen-Dalla Corte; Lamiaa M. Mahmoud; Tatiana S. Moraes; Zhonglin Mou; Jude W. Grosser; Manjul Dutt; Development of Improved Fruit, Vegetable, and Ornamental Crops Using the CRISPR/Cas9 Genome Editing Technique.. Plants 2019, 8, 601, 10.3390/plants8120601.
