Plant diseases annually cause 10–16% yield losses in major crops, prompting urgent innovations. Artificial intelligence (AI) shows an aptitude for automated disease detection and diagnosis utilizing image recognition techniques, with reported accuracies exceeding 95% and surpassing human visual assessment. Forecasting models integrating weather, soil, and crop data enable preemptive interventions by predicting spatial-temporal outbreak risks weeks in advance at 81–95% precision, minimizing pesticide usage. Precision agriculture powered by AI optimizes data-driven, tailored crop protection strategies boosting resilience. Real-time monitoring leveraging AI discerns pre-symptomatic anomalies from plant and environmental data for early alerts. These applications highlight AI’s proficiency in illuminating opaque disease patterns within increasingly complex agricultural data.
- artificial intelligence
- phytopathology
- emerging disease
- climate change
- control diseases
1. Overview of Phytopathology
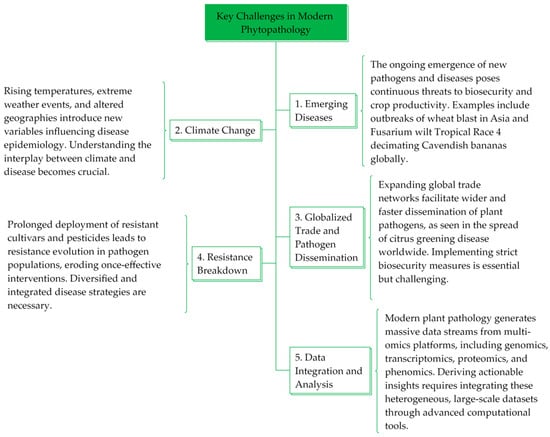
2. Role of Technology in Phytopathology
2.1. Advent of Emerging Technologies in Agriculture
2.2. Need for Advanced Data-Driven Solutions
3. Applications of AI in Phytopathology
Artificial intelligence (AI) is transforming approaches in phytopathology, catalyzing innovations in understanding, managing, and mitigating plant diseases. AI’s capacity to analyze vast datasets reveals subtle correlations in plant–pathogen interactions, granting key insights for disease control [16][36]. This section surveys prominent applications of AI across major facets of phytopathology.3.1. Disease Detection and Diagnosis
Artificial intelligence (AI) enables rapid and precise disease detection and diagnosis, overcoming the limitations of techniques reliant on visual inspection. Numerous studies demonstrate the efficacy of AI in accurately diagnosing complex diseases. In an early example, Ramcharan et al. [14][34] applied deep learning techniques for detecting and diagnosing cassava diseases through image analysis. Using a convolutional neural network, they achieved diagnostic accuracy above 90%, demonstrating deep learning’s effectiveness in identifying various cassava diseases. This approach not only surpassed traditional methods in terms of accuracy and speed but also enabled the implementation of these models on mobile devices, facilitating diagnosis in the field. Similarly, Fuentes et al. [112][37] implemented three artificial intelligence architectures—Faster R-CNN, SSD, and R-FCN—to detect and diagnose diseases and pests in tomato plants. These architectures fall within the broader context of convolutional neural networks (CNNs), which are particularly suited for image recognition tasks due to their ability to learn spatial hierarchies of features from input images. The application of these models in the study marked a significant advancement in the application of CNNs in image recognition tasks since their proposal in the 1990s. The authors of [112][37] used images captured by cameras at various resolutions, both of healthy plants and plants with symptoms. With these images, they trained the artificial processing models, which significantly improved the accuracy in disease and pest recognition and reduced false positives during the training phase. This systematic approach allowed the AI system to effectively recognize nine different types of diseases and pests in tomato plants, demonstrating the capability of these models to handle complex environmental variables present in a plant’s surroundings. Following in the footsteps of these works, but not focused on a specific plant species, Sladojevic et al. [113][38] also used deep convolutional neural networks (CNNs), training the artificial model with an extensive database, which allowed it to distinguish between different types of diseases in the leaves of various genera and species. The novelty and advancement of the developed model lie in its simplicity, where healthy leaves and background images are aligned with other classes, allowing the model to distinguish between diseased and healthy leaves or their surroundings using deep CNNs. The experimental results showed an accuracy of between 91% and 99% in separate class tests and an overall accuracy of 95.8% in the trained model. These studies are a clear example of CNNs’ ability to handle the complexity of visual data and improve the accuracy of automated diagnosis [113][38]. Recent studies have continued to demonstrate AI’s potential in plant disease detection and diagnosis using more modern, precise, and powerful models thanks to the development of new AI capabilities. In 2022, Arinichev [114][39] explored the use of artificial intelligence technologies for diagnosing fungal diseases in cereals, specifically in wheat and rice, through methods of vision and automated recognition. This analysis revealed that artificial neural networks have the capability to detect and classify disease patterns, such as yellow spots, yellow and brown rust, and brown spots, with classification metrics ranging between 0.95 and 0.99. To advance in this line of research, Arinichev examined four well-established and relatively light convolutional neural network (CNN) architectures, namely, GoogleNet, ResNet-18, SqueezeNet-1.0, and DenseNet-121, with the DenseNet-121 model particularly standing out for its optimal combination of high precision and operational efficiency. Characterized by a relatively low number of parameters and a file size suitable for mobile devices, this model achieved exceptionally high classification accuracy, surpassing the other evaluated models. Similar to previous research, such as that of Ramcharan et al., the implementation of a light neural network like DenseNet-121 facilitated its application in the field on mobile devices, allowing for quick and accurate diagnostics [114][39]. In the case of the study carried out by Feng et al. [115][40], the authors developed a convolutional neural network model for potato late blight detection method using deep learning, with high accuracy and fast inference speed, using a dataset of potato leaf disease images in single and complex backgrounds. Feng et al. used the ShuffleNetV2 2× model, characterized by its high classification accuracy, while also having a larger parameter scale and memory space compared to other models with equal accuracy. The authors improved the model through strategies that included introducing an attention module, reducing network depth, and minimizing the number of 1×1 convolutions. This resulted in an enhancement of classification accuracy while simultaneously maintaining efficient inference speed on CPUs in the devices used for its application. In the same line of work, Bracino et al. [116][41] carried out a study focus on the non-destructive classification of paddy rice leaf diseases using deep learning algorithms such as EfficientNet-b0, MobileNet-v2, and Places365-GoogLeNet. They aim to identify whether the rice paddy leaf is normal or infected with various diseases including bacterial leaf blight (BLB), bacterial leaf streaks (BLS), bacterial panicle blight (BPB), heart, downy mildew, hispa, or rice tungro disease (RTD). Of the models used, EfficientNet-b0 was identified as the most effective, achieving an average accuracy of 97.74%. This model is distinguished by its focus on maximizing efficiency, optimally balancing network depth, width, and the resolution of input images through a compound scaling technique, resulting in superior performance with minimal memory requirements and floating-point operations per second (FLOPS). This efficiency and precision capability distinguish it from the model used by Feng et al., the ShuffleNetV2 2×, which, although highly precise, focuses on improving inference speed and reducing parameter size through the introduction of an attention module and the optimization of the network architecture. Bracino et al.’s significant contribution lies in providing a precise and non-destructive diagnostic method for rice diseases, thereby supporting the prevention of product loss and improving crop quality through the application of advanced and efficient AI technologies. A deep convolutional neural network model was also developed by Jouini et al. [117][42] to detect wheat leaf rust. The authors advanced the application of a CNN by developing a model specifically designed for the detection of wheat leaf diseases using hyperspectral images, achieving an impressive testing accuracy of 94%. This study showed the feasibility of real-time disease detection in wheat, a critical advancement for resource-constrained environments where timely and effective disease management is vital [117][42]. In a related study, Zhou et al. [118][43] introduced a novel spectral feature pseudo-graph-based residual network (SFPGRN) for the spectral analysis of plant diseases. Their method innovatively constructs a residual network model using a characteristic surface derived from natural neighborhood interpolation based on preprocessed near-infrared spectral reflection signals and first-order differential spectral index, achieving a classification accuracy of 93.21% on a dataset of apple leaf diseases and insect pests [118][43]. Complementing these developments, Shi et al. [93][44] introduced a novel fast Fourier convolutional deep neural network (FFCDNN) designed for the accurate and interpretable detection of wheat yellow rust and nitrogen deficiency from Sentinel-2 time series data. The FFCDNN model stands out for its innovative use of a fast Fourier convolutional block and a capsule feature encoder, significantly enhancing computing efficiency and model interpretability. This approach not only achieves high classification accuracy but also provides insights into the host–stress interaction, marking a significant advancement over previous studies by integrating spatial-temporal information for global feature extraction [93][44]. In recent times, the research group of Hassan et al. [119][45] introduced a groundbreaking CNN architecture for plant disease identification, leveraging inception layers and residual connections to enhance feature extraction, while employing depth wise separable convolution to significantly reduce computational complexity. This model is distinct in its ability to achieve high accuracy across various plant disease datasets with a markedly lower parameter count, illustrating a significant advancement in AI’s application to phytopathology. By optimizing the model to require fewer computational resources, this work facilitates the deployment of AI technologies on devices with limited processing capabilities, making sophisticated disease diagnosis tools more accessible to a broader range of users and applications [119][45]. A new evolution of CNN is the Siamese convolutional neural network (SNN). Narain et al. [120][46] introduced an enhanced approach to detection systems by implementing a SNN for identifying diseases in tomato leaves. Siamese neural networks stand out from conventional CNN models due to their unique structure, designed to learn to differentiate between pairs of inputs, making them exceptionally suitable for comparison and differentiation tasks. By evaluating similarities or differences between pairs of images, SNNs can offer notable accuracy in disease classification, often overcoming challenges faced by traditional CNNs in terms of intraclass variability and the scarcity of labeled data (Figure 32). In this work, Narain et al. developed a customized SNN by training with a specially collected dataset of 155 tomato leaf images, and the system demonstrated high efficacy, achieving an accuracy of 83.749% in training and 80.4% in testing. This improvement in disease classification represents a significant advancement over more classic CNN models. The implementation of Siamese networks signifies an optimization in the accuracy and efficiency of disease detection in crops, allowing for the application of appropriate management measures more quickly and accurately by providing a more robust and adaptable mechanism for recognizing complex patterns associated with various plant diseases [120][46].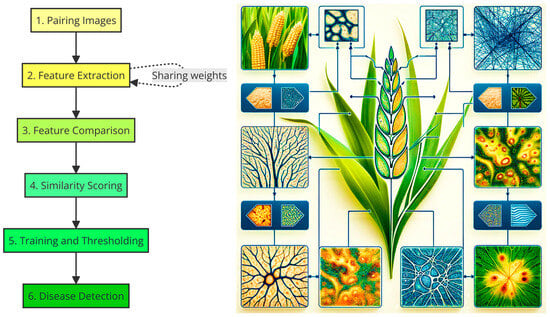
3.2. Advancements in Plant Disease Propagation Modeling
The field of plant disease propagation modeling has witnessed transformative growth through the incorporation of artificial intelligence (AI) and machine learning techniques, opening new vistas in pathogen prediction and management. A pivotal approach in this field is the application of machine learning models for disease prediction based on symptoms and environmental data. In this context, the existing scientific literature encompasses a variety of meticulously developed strategies that significantly contribute to the advancement of predictive model development in the field of plant pathology. During 2022 several studies were published to apply different algorithms and predictive models using AI. For example, a very interesting work is that published by Garrett et al. [39][27] in which the authors utilized Random Forest and Support Vector Machines (SVMs) to analyze the complex interplay between climate change and pathogen emergence. Random Forest is an ensemble learning method for classification, regression, and other tasks. It operates by constructing a multitude of decision trees during training for more accurate and robust predictions (Figure 43). SVMs, in contrast, are powerful supervised machine learning models used for classification and regression challenges, effectively handling high-dimensional spaces and complex datasets. Employing these algorithms, Garrett et al. aimed to capture and model the nuanced relationships between environmental factors and the likelihood of pathogen spread, underscoring the potential of AI to offer predictive insights into plant disease dynamics influenced by climate variables. Their methodology showcases the strengths of combining multiple AI approaches to enhance predictive accuracy and provide actionable insights into pathogen management strategies [39][27]. These modeling approaches were similarly utilized by Otero et al. [122][48], who delved into the creation of data-driven predictive models utilizing artificial intelligence to anticipate the occurrence of Plasmopara viticola and Uncinula necator in the viticultural regions of Southern Europe. Otero et al. employed a variety of machine and deep learning algorithms, including Logistic Regression, Decision Trees, Random Forest, Gradient Boosting, K-Nearest Neighbors, Naïve Bayes, Support Vector Machines, and Deep Neural Networks. Logistic Regression provides a probabilistic approach for binary outcomes, making it suitable for disease presence predictions. Decision Trees offer clear, interpretable decisions. Random Forest improves on Decision Trees by combining multiple trees to reduce overfitting. Gradient Boosting sequentially corrects errors, enhancing model performance. K-Nearest Neighbors classifies based on the majority vote of nearest data points, offering simplicity and effectiveness. Naïve Bayes, based on Bayes’ theorem, excels in classification with an assumption of feature independence. Support Vector Machines efficiently handle high-dimensional data, optimizing margins between classes for clear decision boundaries. Notably, the models employed by Otero et al. achieved over 90% accuracy for infection risk and over 80% for treatment recommendations, highlighting the potential of AI in enhancing disease management strategies in vineyards across Southern Europe [122][48].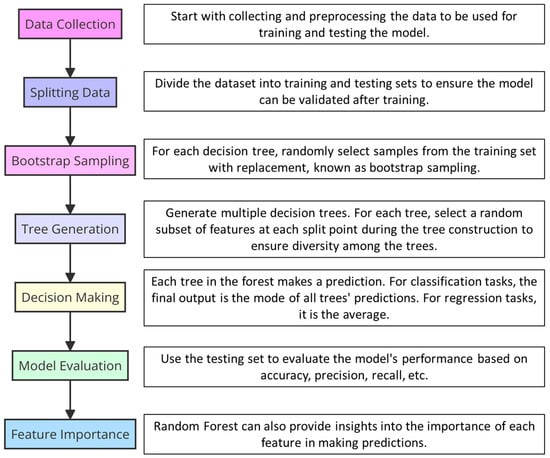
3.3. Comprehensive Evaluation and Prospects of AI Technologies in Phytopathological Applications
In the rapidly evolving domain of AI-assisted plant disease management, the integration of pretrained models, such as ResNet-18 and ResNet-50, marks a significant leap towards refining disease detection and diagnostic accuracy. These models, part of the Residual Networks (ResNets) introduced to mitigate the vanishing gradient problem in deep convolutional neural networks (CNNs), incorporate “shortcut connections” that allow gradients to flow through the network without undergoing non-linear transformations, thereby facilitating the training of much deeper networks. ResNet-50, a 50-layer CNN comprising 48 convolutional layers, one MaxPool layer, and one average pool layer, employs a “bottleneck” design in each residual block to reduce the number of parameters and accelerate layer training. This bottleneck design, featuring a stack of three layers instead of two, utilizes 1 × 1 convolutions to compress and then expand the number of channels, significantly lowering computational complexity while maintaining or enhancing model performance [129][55]. Originally trained on the expansive ImageNet dataset, these models exhibit exceptional prowess in feature recognition, offering tailored solutions for the nuanced challenges of phytopathology. Their capability to discern complex image characteristics with remarkable precision positions them as indispensable tools for identifying plant pathologies, often surpassing traditional visual inspection methods with accuracies ranging between 95% and 97%. Furthermore, the application of these pretrained models extends beyond disease identification to encompass broader spatial analyses, as evidenced by their deployment within the ArcGIS ecosystem for tasks like land cover classification and aerial feature extraction, underscoring their potential to revolutionize the monitoring and management of plant health on a large scale [130][56]. The comparative analysis of AI models, including CNNs, YOLOv5, and MobileNet, illuminates the diverse applicability and efficacy of these technologies in phytopathology. Each model, with its unique strengths (be it CNNs for their image processing capabilities, YOLOv5 for its rapid processing speed facilitating timely interventions, or MobileNet for offering an efficient solution on low-power devices), advances our capacity to manage plant disease spread through predictive analyses that integrate environmental and symptomatic data. This synthesis not only augments diagnostic precision but also enhances proactive disease management strategies. Nevertheless, the practical application of these models encounters challenges such as the need for image preprocessing and handling unbalanced datasets, propelling the pursuit of technological innovations, especially in the realm of Edge-AI devices. These advancements promise a transformative impact on disease monitoring, enabling more accurate and accessible diagnostics. As AI technologies continue to evolve, alongside breakthroughs in sensor technologies, we are ushered towards a new era of integrated and automated plant disease management. This journey is not without its hurdles, necessitating innovative approaches like transfer learning and the development of multisensorial detection systems to overcome current limitations. The ongoing exploration and refinement of AI models in phytopathology not only pave the way for future research directions but also highlight the pivotal role of AI in crafting sustainable, precision-based solutions for global agricultural challenges.References
- Agrios, G.N. Plant Pathology, 5th ed.; Academic Press: Amsterdam, The Netherlands, 2004; ISBN 9780120445653.
- Strange, R.N.; Scott, P.R. Plant Disease: A Threat to Global Food Security. Annu. Rev. Phytopathol. 2005, 43, 83–116.
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging Infectious Diseases of Plants: Pathogen Pollution, Climate Change and Agrotechnology Drivers. Trends Ecol. Evol. 2004, 19, 535–544.
- Oerke, E.C. Crop Losses to Pests. J. Agric. Sci. 2005, 144, 31–43.
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The Global Burden of Pathogens and Pests on Major Food Crops. Nat. Ecol. Evol. 2019, 3, 430–439.
- Legg, J.P.; Lava Kumar, P.; Makeshkumar, T.; Tripathi, L.; Ferguson, M.; Kanju, E.; Ntawuruhunga, P.; Cuellar, W. Cassava Virus Diseases: Biology, Epidemiology, and Management. Adv. Virus Res. 2015, 91, 85–142.
- Ploetz, R.C. Fusarium Wilt of Banana. Phytopathology 2015, 105, 1512–1521.
- Lucas, J.A.; Hawkins, N.J.; Fraaije, B.A. The Evolution of Fungicide Resistance. Adv. Appl. Microbiol. 2015, 90, 29–92.
- Gonsalves, D. Control Of Papaya Ringspot Virus In Papaya: A Case Study. Annu. Rev. Phytopathol. 1998, 36, 415–452.
- Avelino, J.; Cristancho, M.; Georgiou, S.; Imbach, P.; Aguilar, L.; Bornemann, G.; Läderach, P.; Anzueto, F.; Hruska, A.J.; Morales, C. The Coffee Rust Crises in Colombia and Central America (2008–2013): Impacts, Plausible Causes and Proposed Solutions. Food Secur. 2015, 7, 303–321.
- Gottwald, T.R. Current Epidemiological Understanding of Citrus Huanglongbing*. Annu. Rev. Phytopathol. 2010, 48, 119–139.
- Pethybridge, S.J.; Nelson, S.C. Leaf Doctor: A New Portable Application for Quantifying Plant Disease Severity. Plant Dis. 2015, 99, 1310–1316.
- Mahlein, A.K. Plant Disease Detection by Imaging Sensors—Parallels and Specific Demands for Precision Agriculture and Plant Phenotyping. Plant Dis. 2016, 100, 241–254.
- Ahmad, A.; Hettiarachchi, R.; Khezri, A.; Singh Ahluwalia, B.; Wadduwage, D.N.; Ahmad, R. Highly Sensitive Quantitative Phase Microscopy and Deep Learning Aided with Whole Genome Sequencing for Rapid Detection of Infection and Antimicrobial Resistance. Front. Microbiol. 2023, 14, 1154620.
- Proaño-Cuenca, F.; Espindola, A.S.; Garzon, C.D. Detection of Phytophthora, Pythium, Globisporangium, Hyaloperonospora, and Plasmopara Species in High-Throughput Sequencing Data by In Silico and In Vitro Analysis Using Microbe Finder (MiFi). PhytoFrontiers 2023, 3, 124–136.
- Kemen, E. Microbe–Microbe Interactions Determine Oomycete and Fungal Host Colonization. Curr. Opin. Plant Biol. 2014, 20, 75–81.
- Joshi, A.; Song, H.G.; Yang, S.Y.; Lee, J.H. Integrated Molecular and Bioinformatics Approaches for Disease-Related Genes in Plants. Plants 2023, 12, 2454.
- Venbrux, M.; Crauwels, S.; Rediers, H. Current and Emerging Trends in Techniques for Plant Pathogen Detection. Front. Plant Sci. 2023, 14, 1120968.
- Qin, J.; Monje, O.; Nugent, M.R.; Finn, J.R.; O’Rourke, A.E.; Wilson, K.D.; Fritsche, R.F.; Baek, I.; Chan, D.E.; Kim, M.S. A Hyperspectral Plant Health Monitoring System for Space Crop Production. Front. Plant Sci. 2023, 14, 1133505.
- Schirrmann, M. Advances in Remote Sensing Technologies for Assessing Crop Health. In Advances in Sensor Technology for Sustainable Crop Production; Burleigh Dodds Science Publishing Limited: Cambridge, UK, 2023; pp. 43–64. ISBN 9781786769770.
- Abbas, A.; Zhang, Z.; Zheng, H.; Alami, M.M.; Alrefaei, A.F.; Abbas, Q.; Naqvi, S.A.H.; Rao, M.J.; Mosa, W.F.A.; Abbas, Q.; et al. Drones in Plant Disease Assessment, Efficient Monitoring, and Detection: A Way Forward to Smart Agriculture. Agronomy 2023, 13, 1524.
- Hu, H.; Wang, N.; Liao, J.; Tovar-Lopez, F.J. Recent Progress in Micro- and Nanotechnology-Enabled Sensors for Biomedical and Environmental Challenges. Sensors 2023, 23, 5406.
- Arshad, F.; Deliorman, M.; Sukumar, P.; Qasaimeh, M.A.; Olarve, J.S.; Santos, G.N.; Bansal, V.; Ahmed, M.U. Recent Developments and Applications of Nanomaterial-Based Lab-on-a-Chip Devices for Sustainable Agri-Food Industries. Trends Food Sci. Technol. 2023, 136, 145–158.
- Kamilaris, A.; Kartakoullis, A.; Prenafeta-Boldú, F.X. A Review on the Practice of Big Data Analysis in Agriculture. Comput. Electron. Agric. 2017, 143, 23–37.
- Glass, C.R.; Gonzalez, F.J.E. Developing of New Technologies Driving Advances in Precision Agriculture to Optimise Inputs and Reduce Environmental Footprint. C3-BIOECONOMY Circ. Sustain. Bioeconomy 2022, 69–75.
- Kotpalliwar, P.; Barmate, M.; Satpute, P.; Manapure, D.; Hassan, M.; Student, U.G.; Prof, A. Agro Analysis System for Precision Agriculture. Int. J. Res. Appl. Sci. Eng. Technol. (IJRASET) 2023, 11, 960–963.
- Garrett, K.A.; Bebber, D.P.; Etherton, B.A.; Gold, K.M.; Sulá, A.I.P.; Selvaraj, M.G. Climate Change Effects on Pathogen Emergence: Artificial Intelligence to Translate Big Data for Mitigation. Annu. Rev. Phytopathol. 2022, 60, 357–378.
- Tatineni, S.; Hein, G.L. Plant Viruses of Agricultural Importance: Current and Future Perspectives of Virus Disease Management Strategies. Phytopathology 2023, 113, 117–141.
- Collinge, D.B.; Jensen, D.F.; Rabiey, M.; Sarrocco, S.; Shaw, M.W.; Shaw, R.H. Biological Control of Plant Diseases—What Has Been Achieved and What Is the Direction? Plant Pathol. 2022, 71, 1024–1047.
- Ferguson, M.; Hsu, C.K.; Grim, C.; Kauffman, M.; Jarvis, K.; Pettengill, J.B.; Babu, U.S.; Harrison, L.M.; Li, B.; Hayford, A.; et al. A Longitudinal Study to Examine the Influence of Farming Practices and Environmental Factors on Pathogen Prevalence Using Structural Equation Modeling. Front. Microbiol. 2023, 14, 1141043.
- Miller, S.A.; Ferreira, J.P.; Lejeune, J.T. Antimicrobial Use and Resistance in Plant Agriculture: A One Health Perspective. Agriculture 2022, 12, 289.
- Banerjee, S.; Mondal, A.C. An Intelligent Approach to Reducing Plant Disease and Enhancing Productivity Using Machine Learning. Int. J. Recent Innov. Trends Comput. Commun. 2023, 11, 250–262.
- Singh, A.; Ganapathysubramanian, B.; Singh, A.K.; Sarkar, S. Machine Learning for High-Throughput Stress Phenotyping in Plants. Trends Plant Sci. 2016, 21, 110–124.
- Ramcharan, A.; Baranowski, K.; McCloskey, P.; Ahmed, B.; Legg, J.; Hughes, D.P. Deep Learning for Image-Based Cassava Disease Detection. Front. Plant Sci. 2017, 8, 1852.
- Tzachor, A.; Devare, M.; King, B.; Avin, S.; Héigeartaigh, S.Ó. Responsible Artificial Intelligence in Agriculture Requires Systemic Understanding of Risks and Externalities. Nat. Mach. Intell. 2022, 4, 104–109.
- Zhao, L.; Walkowiak, S.; Fernando, W.G.D. Artificial Intelligence: A Promising Tool in Exploring the Phytomicrobiome in Managing Disease and Promoting Plant Health. Plants 2023, 12, 1852.
- Fuentes, A.; Yoon, S.; Kim, S.C.; Park, D.S. A Robust Deep-Learning-Based Detector for Real-Time Tomato Plant Diseases and Pests Recognition. Sensors 2017, 17, 2022.
- Sladojevic, S.; Arsenovic, M.; Anderla, A.; Culibrk, D.; Stefanovic, D. Deep Neural Networks Based Recognition of Plant Diseases by Leaf Image Classification. Comput. Intell. Neurosci. 2016, 2016, 3289801.
- Arinichev, I.V. Using Digital Intelligent Technologies for the Diagnosis of Cereals Diseases in the Kuban. Agrar. Sci. J. 2022, 5, 70–73.
- Feng, J.; Hou, B.; Yu, C.; Yang, H.; Wang, C.; Shi, X.; Hu, Y.; Feng, J.; Hou, B.; Yu, C.; et al. Research and Validation of Potato Late Blight Detection Method Based on Deep Learning. Agronomy 2023, 13, 1659.
- Bracino, A.A.; Evangelista, D.G.D.; Concepcion, R.S.; Dadios, E.P.; Vicerra, R.R.P. Non-Destructive Classification of Paddy Rice Leaf Disease Infected by Bacterial and Fungal Species Using Vision-Based Deep Learning. J. Adv. Comput. Intell. Intell. Inform. 2023, 27, 333–339.
- Jouini, O.; Sethom, K.; Bouallegue, R. Wheat Leaf Disease Detection Using CNN in Smart Agriculture. In Proceedings of the 2023 International Wireless Communications and Mobile Computing, IWCMC 2023, Marrakesh, Morocco, 19–23 June 2023; Institute of Electrical and Electronics Engineers Inc.: New York, NY, USA, 2023; pp. 1660–1665.
- Zhou, Y.; Pan, Z.; Liu, Z.; He, T.; Yan, J. SFPGRN: Spectral Detection Method of Plant Diseases Based on Deep Learning. SPIE 2023, 12717, 127171F.
- Shi, Y.; Han, L.; González-Moreno, P.; Dancey, D.; Huang, W.; Zhang, Z.; Liu, Y.; Huang, M.; Miao, H.; Dai, M. A Fast Fourier Convolutional Deep Neural Network for Accurate and Explainable Discrimination of Wheat Yellow Rust and Nitrogen Deficiency from Sentinel-2 Time Series Data. Front. Plant Sci. 2023, 14, 1250844.
- Hassan, S.M.; Maji, A.K. Plant Disease Identification Using a Novel Convolutional Neural Network. IEEE Access 2022, 10, 5390–5401.
- Narain, T.; Sahu, P.; Singh, A.P. Plant Disease Classification Using Siamese Convolutional Neural Network. Lect. Notes Netw. Syst. 2023, 471, 57–65.
- Tabbakh, A.; Barpanda, S.S. A Deep Features Extraction Model Based on the Transfer Learning Model and Vision Transformer “TLMViT” for Plant Disease Classification. IEEE Access 2023, 11, 45377–45392.
- Otero, M.; Velasquez, L.F.; Basile, B.; Onrubia, J.R.; Pujol, A.J.; Pijuan, J. Data Driven Predictive Models Based on Artificial Intelligence to Anticipate the Presence of Plasmopara Viticola and Uncinula Necator in Southern European Winegrowing Regions. Front. Artif. Intell. Appl. 2022, 356, 164–167.
- Lavanya, A.; Krishna, T.M. An AI and Cloud Based Collaborative Platform for PlantDisease Identification, Tracking and Forecasting for Farmers. Int. J. Eng. Technol. Manag. Sci. 2022, 6, 527–537.
- Zen, B.P.; Iqsyahiro Kresna, A.; Fransisca, D.C. Applications for Detecting Plant Diseases Based on Artificial Intelligence. SinkrOn 2022, 7, 2537–2546.
- Marco-Detchart, C.; Carrascosa, C.; Julian, V.; Rincon, J. Robust Multi-Sensor Consensus Plant Disease Detection Using the Choquet Integral. Sensors 2023, 23, 2382.
- Ojo, M.O.; Zahid, A. Improving Deep Learning Classifiers Performance via Preprocessing and Class Imbalance Approaches in a Plant Disease Detection Pipeline. Agronomy 2023, 13, 887.
- Vardhan, J.; Swetha, K.S. Detection of Healthy and Diseased Crops in Drone Captured Images Using Deep Learning. arXiv 2023, arXiv:2305.13490.
- Dagwale, S.S.; Adakane, P. Prediction of Leaf Species & Disease Using Ai for Various Plants. Int. J. Multidiscip. Res. 2023, 5, 23034169.
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778.
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Kai, L.; Li, F.-F. ImageNet: A Large-Scale Hierarchical Image Database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255.
