Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Camila Xu and Version 1 by Vijay Kumar.
Pattern recognition receptors (PRRs) play critical roles in embryonic development, immune homeostasis, neurodevelopment, and neurodegeneration. PRRs are highly conserved germline-encoded proteins that recognize microbe/pathogen-associated molecular patterns (MAMPs or PAMPs) and death/damage-associated molecular patterns (DAMPs); thus, they regulate innate and adaptive immunity and contribute to the pathogenesis of many diseases ranging from infections to cancers.
- PRRs
- cGLRs
- cGAS
- STING
- type 1 IFN
1. Introduction
Pattern recognition receptors (PRRs) play critical roles in embryonic development, immune homeostasis, neurodevelopment, and neurodegeneration. PRRs are highly conserved germline-encoded proteins that recognize microbe/pathogen-associated molecular patterns (MAMPs or PAMPs) and death/damage-associated molecular patterns (DAMPs) [1,2,3][1][2][3]; thus, they regulate innate and adaptive immunity [1,4,5][1][4][5] and contribute to the pathogenesis of many diseases ranging from infections to cancers [5,6,7,8,9,10,11][5][6][7][8][9][10][11]. Traditionally, PRR family members have included toll-like receptors (TLRs), C-type lectin receptors (CLRs), nucleotide-oligomerization domain (NOD)-like receptors (NLRs), absent in melanoma-2-like receptors (ALRs), retinoic acid-inducible gene (RIG)-1-like receptors (RLRs), C-type lectin receptors (CLRs), and different scavenger receptors (SRs) [2,9,12,13,14,15,16,17][2][9][12][13][14][15][16][17]. Cytosolic guanosine monophosphate (GMP)-adenosine monophosphate (AMP) synthase (cGAS)-like receptors (cGLRs) have been recently recognized as new PRRs that potentially interact with other PRRs to regulate immune homeostasis.
2. Emergence of cGLRs (Harboring cGAS and STING) as a Novel Family of PRRs
cGLRs have emerged or evolved as critical PRRs providing defense through different mechanisms in prokaryotes and eukaryotes, including humans. Table 1 shows different cGLRs and their known ligands. The first enzyme responsible for synthesizing cyclic guanosine monophosphate (GMP)-adenosine monophosphate (AMP) (cGAMP) or cyclicdinucleotides (CDNs), dinucleotide cyclase (DncV), or cGAS-DncV-like nucleotidyltransferase (CD-NTase), evolved in bacteria and has been reported in Vibrio cholerae. CD-NTases generating CDNs, like animal cGASs, are very diverse proteins numbering over 6000 to date [18]. In addition to producing CDNs, bacterial CD-NTases can also produce cyclic trinucleotides (CTNs) and linear oligonucleotides [19,20][19][20]. Although DncV lacks primary sequence homology to h-cGAS (synthesizes 2′3′-cGAMP), it synthesizes conventional 3′3′- cGAMP involved in bacterial chemotaxis [21,22][21][22]. Bacteria encode thousands of cGAS/DncV-like NTases (CD-NTases) to control their highly divergent anti-phage defense system. Furthermore, DncV NTases, cGAS homologs, and 2′-5′-oligoadenylate synthase 1 (OAS1, having aspartic acid (Asp) in place of E225) have five highly conserved active sites [22]. The CD-NTases of the cyclic oligonucleotide-based antiphage signaling system (CBASS, analogous to the cGAS/STING signaling pathway of metazoans) that are activated by the binding of a folded fragment of RNA share distant homology with the cGAS [23,24,25,26,27][23][24][25][26][27]. Furthermore, CBASS-activating bacteriophage RNA (cabRNA), via binding to the surface of positively charged CdnEO3 cyclase (a bacterial cGAS), promotes cGAMP synthesis to activate the CBASS-mediated immune response [28].Thus, the CBASS in the bacteria producing cGAMP serves as a defense system against bacteriophages [29]. The bacterial cGAS or CBASS comprises a four-gene operon that encodes bacterial cGAS, which is associated with phospholipase, two enzymes with the eukaryotic-like domains E1 and E2, and a Janus kinase-binding protein (JAB) domain, which provides resistance against several bacteriophages. cGAMP production activates phospholipases to disintegrate membranes and bacterial cell death before the completion of the bacteriophage life cycle or the production of mature bacteriophage [29].Table 1. Different cGLRs (including mammalian cGAS), their ligands, cleaved products (cyclic dinucleotides, and CDNs such as 2′3′-cGAMP and/or 3′3′-cGAMP) activating STING and downstream signaling molecules throughout the animal kingdom from metazoans to mammals. Kindly see the article text and referenced articles for details.
| Species | cGLR | Ligand/Product | STING | IKKε | TBK1 | NF-κB | IRF3 | IRF7 |
|---|---|---|---|---|---|---|---|---|
| Homo sapiens/Human | cGAS | dsDNA/2′3′-cGAMP | + | + | + | + | + | + |
| Mus musculus/Laboratory mouse | cGAS | dsDNA/2′3′-cGAMP | + | + | + | + | + | + |
| Gallus gallus/Red junglefowl | cGAS | dsDNA/2′3′-cGAMP | + | + | + | + | - | + |
| Xenopus tropicalis/Western clawed frog | cGAS | dsDNA/2′3′-cGAMP | + (without CTT domain) | + | + | + | + | + |
| Branchiostoma floridae/Florida lancelet | 2 cGAS homologs | dsDNA/2′3′-cGAMP | +, 2 STING candidates (STING-1 and STING-2, | + | + | + | - | - |
| Danio rerio/Zebrafish | 2 cGAS | Cytosolic dsDNA/2′3′-cGAMP | + | + | + | + | + | + |
| Strongylocentrotus purpuratus/Pacific purple sea urchin | ? | ? | - | + | + | + | - | - |
| Ceanorhabidits elegans/Roundworm | - | - | - | + | - | - | - | - |
| Drosophila melanogaster/Common fruit fly | Dm-cGLR1 and Dm-cGLR2 | dsRNA/3′2′-cGAMP and 2′3′-cGAMP | + | - | + | + | - | - |
| Tribolium castaneium/Red flour beetle | Tc-cGLR | dsRNA/2′3′-cGAMP | + | - | + | + | - | - |
| Microplitis demolitor/Wasp | Md-cGLR | dsRNA/2′3′-cGAMP | + | - | + | + | - | - |
| Frankliniella occidentalis/Western flower thrip | Fo-cGLR | dsRNA/2′3′-cGAMP | + | ? | + | + | - | - |
| Nicrophorus vespilloides/Common sexton beetle | Nves-cGLR | dsRNA/2′3′-cGAMP | + | ? | + | + | - | - |
| Aethina tumida/Small hive beetle | At-cGLR | dsRNA/2′3′-cGAMP | + | ? | + | + | - | - |
| Asbolus verrucosus/Blue death feigning beetle | Av-cGLR | dsRNA/2′3′-cGAMP | + | ? | + | + | - | - |
| Trichogramma pretiosum/Wasp | Tp-cGLR | Unknown/2′3′-cGAMP | + | - | + | + | - | - |
| Chlamydophila felis/Cat flea | Cf-cGLR | Unknown/2′3′-cGAMP | ? | ? | + | + | ? | ? |
| Pocilloporidae damicornis/Cauliflower coral | Pd-cGLR | Unknown/2′3′-cGAMP | + | - | + | + | - | ? |
| Crassostrea gigas/Pacific oyster | Cg-cGLR1 | dsDNA/2′3′-cUA | + | + | + | + | -, but have cgIRF1 | -, but have cgIRF8 |
| Crassostrea virginica/Eastern oyster | Cv-cGLR1 | dsDNA/2′3′-cUA | + | + | + | + | -, but have cgIRF1 | -, but have cgIRF8 |
| Cv-cGLR2 | Unknown/Unknown | |||||||
| Sp-cGLR1 | dsRNA/3′3′-cUA | |||||||
| Stylophora pistillata/Stony coral | Sp-cGLR2 | Unknown/2′3′-cGAMP | + | - | ? | + | ? | ? |
| Sp-cGLR3 | dsRNA/3′3′-cAA | |||||||
| Amphimedon queenslandica/Sponge | ? | ? | - | - | + | + | - | - |
| Exaiptasia pallida/Glass anemone | Ep-cGLR | Unknown/2′3′-cGAMP | + | - | + | + | - | - |
| Monosiga brevicollis/Choanoflagellate | Mb-cGAS | dsDNA/3′3′-cGAMP | + | - | - | - | - | - |
| Nematostella vectensis/Starlet Sea anemone | Nv-cGAS or nvA7SFB5.1 | dsDNA/3′3′-cGAMP | + | - | + | + | - | - |
| Hydra magnipapillata/Hydra vulgaris | Hv-cGLR | dsRNA/2′3′-cGAMP | +, 3 STING candidates (STING-1, STING-2, and STING-3) | + | - | + | - | - |
+ = present; - = absent; ? = not known/unsure; cUA, cyclic UMP-AMP; cAA, cyclic diAMP.
Phylogenetic studies have indicated that animal cGAS, or chromosome 6 open reading frame 150 (C6orf150), and STING date back to the origin of choanoflagellate (nearest free-living unicellular and colonial flagellates related to metazoans) called Monosiga brevicollis [22]. However, cGAS and STING proteins have substantially different origins, as STINGs arose via convergent domain shuffling in bacteria and eukaryotes, while cGAS homologs or eukaryotic CD-NTases arose due to multiple horizontal gene transfer (HGT) events from bacteria to eukaryotes [18]. Notably, cGAS homologs of invertebrates may not always recognize dsDNA as a PAMP/MAMP and DAMP. Thus, within metazoans, homologs of both cGAS and STING are present as early as in cnidarians including Nematostella vectensis and Hydra magnipapillata [22]. cGAS and STING are present in all Drosophila species, many non-Drosophila arthropods, and almost all chordates except torafugu Takifugu rubripes. Interestingly, cGAS and STING are absent in the flatworm Schistosoma mansoni and nematodes such as Caenorhabditis elegans [22,24][22][24]. Furthermore, they are present in H. magnipapillata (Hydra), Tribolium. Castaneum (Red Flour Beetle), Drosophila virilis, Drosophila persimilis, Drosophila pseudoobscura, cephalochordate Brachiostoma floridae (Florida lancelet), and Danio rerio (zebrafish), each of which has two cGAS homologs. Also, H. magnipapillata contains three STING candidates (STING-1, STING-2, and STING-3), while B. floridae harbors two STING candidates (STING-1 and STING-2) (Table 1) [22].
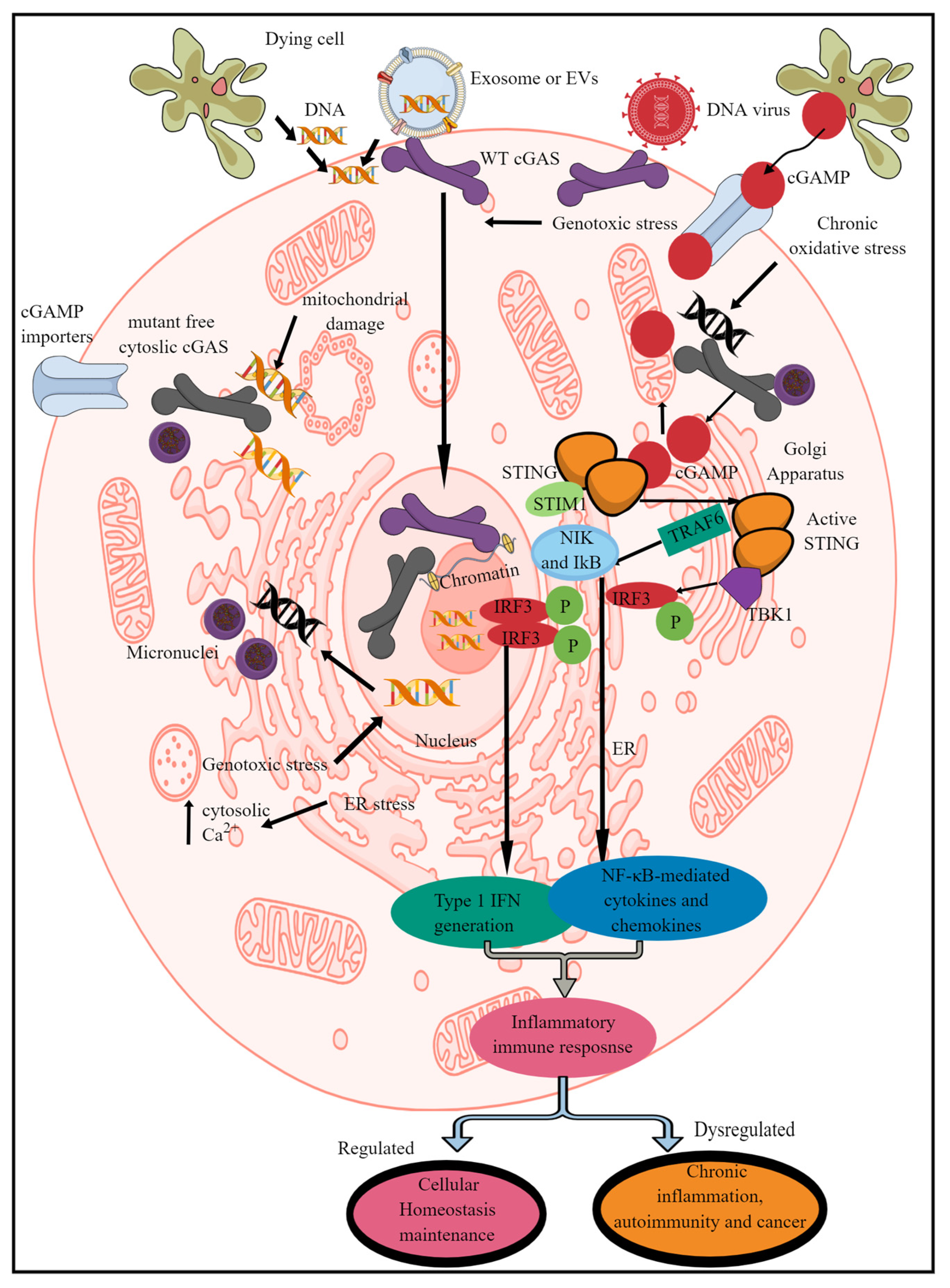
cGAS/STING/TBK1 axis-dependent type 1 IFN generation exists in mammals (humans and mice), fish (D. rerio and Oryzias latipes), insects, (Drosophila spp., wasps), molluscs, (oysters), and cnidarians (sea anemone, Hydra, corals) (Table 1) [24]. The cGAS or C6orf150 or male abnormal 21 (Mab21) domain-containing protein 1 recognizes self- and pathogen-derived cytosolic DNA to activate the innate immune response (Figure 1) [30]. Human cGAS (h-cGAS) comprises an unstructured and poorly conserved N terminus (amino acid (AA) residues 1–160) and a highly conserved C terminus (160–513). The C terminus consists of a conserved nucleotidyltransferase (NTase) core domain (160–330) and a Mab21 domain (213–513) that contains a zinc (Zn2+)-ribbon structural domain (390–405) [22]. This Zn2+ ribbon domain is conserved among all vertebrate cGAS members and cGLRs, excluding three homologs in Pan paniscus (bonobo), Canis familiaris (dog), and Taeniopygia guttata (zebra finch) [22].
 Mab21 was first recognized as the fate-determining gene for the morphogenesis of the sensory rays, a male-specific sense organ located in the tail and involved in copulation in the C. elegans [31]. The invertebrate Litopenaeus vannamei (Whiteleg Shrimp or King Prawn) Mab21 (LvMab21cp) has a similar expression profile to LvSTING and LvIRF and serves as a human analog of the vertebrate cGAS, which recognizes viruses and generates antiviral immune responses [32]. Mouse cGAS (m-cGAS) also contains the Mab21 domain; however, eukaryotic homologs of m-cGAS are more similar to m-cGAS than mouse Mab21-like proteins [22]. Notably, h-cGAS structurally and enzymatically resembles human OAS1, a template-independent nucleotidyl transferase that activates antiviral innate immunity upon recognizing cytosolic short dsRNA (>18 bp) [33,34,35][33][34][35]. Notably, OAS-like proteins also have an evolutionary prokaryotic or bacterial origin through the development of antiviral defense and are independently acquired by eukaryotes [18]. Furthermore, the Zn2+-ribbon domain of vertebrate cGAS is absent in OAS1, nematode Mab-21, and mammalian Mab-21-like proteins [22]. The presence of the elongated N-terminal (167 AAs) in vertebrate and cephalochordate (B. floridae) cGAS has evolved as an adaptation that is very short in invertebrates (varying from fewer than 7 AAs to 70) [22].
Both h-cGAS and m-cGAS have two DNA binding sites and form a dimeric complex with two dsDNAs [36,37][36][37]. Of the five AA residues that are critical for 2′3′-cGAMP binding, only S434 of h-cGAS is non-conserved in mammals, whereas Y436 is conserved in all species. The three remaining residues (K362, R376, and S378) are wholly conserved in cephalochordates and vertebrates but not in arthropods [22]. The details of conserved and non-conserved AA residues in the dsDNA binding sites of cGAS have been discussed elsewhere [22]. The four non-conserved AA residues (R236, K254, K327, and R353) in vertebrates suggest that only double and triple mutations inhibit cGAS’s ability to stimulate type 1 IFN production [22,36][22][36]. Thus, the NTase system has evolved as a defense system against foreign and delocalized genetic material that enters the cytosol from the mitochondria and nucleus. These findings demonstrate that GLRs are critical family members of PRRs.
More than three thousand cGLRs with complete active sites representing nearly all major animal phyla have been identified [24]. Furthermore, cGLR sequence analysis has delineated specific evolutionary patterns in animal immunity and protein features that are distinct from bacterial CD-NTase anti-phage defense enzymes [24]. Animal cGLRs form a single family of signaling proteins that share more distantly related homologies with bacterial CD-NTase anti-phage defense enzymes and animal OAS1-like proteins [18,24][18][24]. Furthermore, the interaction between STING and CDNs is universally conserved. However, some rare bacteriophages avoid potent innate immune responses by preventing cGAS/STING signaling through major capsid gene mutations in the CBASS [38,39,40][38][39][40]. Even bacteriophages escaping the bacterial cGLR or CBASS generate longer cabRNA that prevents the recognition and activation of CdnEO3.
STING-mediated CDN recognition also originated in bacteria as a defense mechanism against bacteriophages. Bacterial STING (b-STING) forms protein filaments to drive oligomerization of Toll/Interleukin 1 receptor (TIR) effector domains and rapid NAD+ cleavage [23]. STINGs and bacterial-like STING (bl-STINGs, present in microeukaryotes clustered between bacterial and metazoan sequences) share similarities in their domains [18]. Most new eukaryotic bl-STINGs have four N-terminal alpha helices as seen in human STING (h-STING). Furthermore, bacterial transmembrane domain -STINGs (TM-STINGS) have two N-terminal alpha helices and are more similar to bacterial Toll/Interleukin 1 receptor (TIR)-STING or TIR-STING, indicating eukaryotes and bacteria independently converged on a typical TM-STING domain architecture through domain shuffling [18]. Notably, TIR domains of oyster TIR-STING are related to animal TIRs and differ from the TIR domains of bacterial TIR-STINGs. Eukaryotic TIR-STINGs are rare, supporting the hypothesis that this protein originated from recent animal convergence or convergent domain shuffling [18,41][18][41]. Therefore, TM-STING and TIR-STING proteins have evolved due to two independent convergent evolution processes, and bacteria and eukaryotes have used similar proteins by reusing ancient protein domains.
The Drosophila STING (DmSTING or CG1667) is an ortholog to the vertebrate STING. DmSTING has evolutionarily conserved antibacterial (Listeria monocytogenes) and antiviral (positive-strand RNA Drosophila C virus and other DNA viruses) activity that involves the downstream activation of immune deficiency (IMD) and the Drosophila inhibitory kinase kinase β (dIKKβ) pathway and the subsequent cleavage of Relish (an NF-κB homolog) to generate antimicrobial peptides (AMPs) without the involvement of its cGAS ortholog CG7194. CG7194 lacks both the Zn2+-ribbon domain and a positively charged N terminus which are critical for DNA binding [42,43,44,45,46][42][43][44][45][46]. Furthermore, DmSTING transferred to mammalian cells induces an NF-κB-dependent immune response. Hence, Drosophila cGLRs, especially cGLR1, sense cytosolic dsRNA to generate the 3′2′-cGAMP that DmSTING recognizes to initiate the downstream innate immune response through activating Relish [47,48][47][48].
Drosophila cGLR2 generates 2′3′-cGAMP and 3′2′-cGAMP through an unknown target to generate a protective antimicrobial immune response [48]. The loss of DmSTING in Parkin RBR E3 ubiquitin-protein ligase KO (parkin−/−) flies rescues thorax muscle defects, climbing ability, and disrupted mitochondrial morphology of their flight muscles, indicating a critical noncanonical role of DmSTING in cellular and organismal responses to mitochondria stress [49]. Flies lacking parkin and DmSTING exhibit PTEN-induced kinase 1 (PINK1) activation to suppress cell death. Parkin, PINK1, and mitochondrial fidelity play critical roles in the pathogenesis of Parkinson’s disease (PD) in humans [50,51,52][50][51][52]; therefore, it would be interesting to see if a similar h-STING-Parkin-PINK-1 axis exists in human patients with PD. Thus, the cGAS/STING signaling-mediated immune defense mechanism is evolutionarily very ancient, with its origin dating back to prokaryotes/bacteria fighting against invading viruses. Eukaryotes, including humans, have acquired it via convergent domain shuffling and multi-HGT to guard against viruses and self-derived cytosolic DNAs.
Mab21 was first recognized as the fate-determining gene for the morphogenesis of the sensory rays, a male-specific sense organ located in the tail and involved in copulation in the C. elegans [31]. The invertebrate Litopenaeus vannamei (Whiteleg Shrimp or King Prawn) Mab21 (LvMab21cp) has a similar expression profile to LvSTING and LvIRF and serves as a human analog of the vertebrate cGAS, which recognizes viruses and generates antiviral immune responses [32]. Mouse cGAS (m-cGAS) also contains the Mab21 domain; however, eukaryotic homologs of m-cGAS are more similar to m-cGAS than mouse Mab21-like proteins [22]. Notably, h-cGAS structurally and enzymatically resembles human OAS1, a template-independent nucleotidyl transferase that activates antiviral innate immunity upon recognizing cytosolic short dsRNA (>18 bp) [33,34,35][33][34][35]. Notably, OAS-like proteins also have an evolutionary prokaryotic or bacterial origin through the development of antiviral defense and are independently acquired by eukaryotes [18]. Furthermore, the Zn2+-ribbon domain of vertebrate cGAS is absent in OAS1, nematode Mab-21, and mammalian Mab-21-like proteins [22]. The presence of the elongated N-terminal (167 AAs) in vertebrate and cephalochordate (B. floridae) cGAS has evolved as an adaptation that is very short in invertebrates (varying from fewer than 7 AAs to 70) [22].
Both h-cGAS and m-cGAS have two DNA binding sites and form a dimeric complex with two dsDNAs [36,37][36][37]. Of the five AA residues that are critical for 2′3′-cGAMP binding, only S434 of h-cGAS is non-conserved in mammals, whereas Y436 is conserved in all species. The three remaining residues (K362, R376, and S378) are wholly conserved in cephalochordates and vertebrates but not in arthropods [22]. The details of conserved and non-conserved AA residues in the dsDNA binding sites of cGAS have been discussed elsewhere [22]. The four non-conserved AA residues (R236, K254, K327, and R353) in vertebrates suggest that only double and triple mutations inhibit cGAS’s ability to stimulate type 1 IFN production [22,36][22][36]. Thus, the NTase system has evolved as a defense system against foreign and delocalized genetic material that enters the cytosol from the mitochondria and nucleus. These findings demonstrate that GLRs are critical family members of PRRs.
More than three thousand cGLRs with complete active sites representing nearly all major animal phyla have been identified [24]. Furthermore, cGLR sequence analysis has delineated specific evolutionary patterns in animal immunity and protein features that are distinct from bacterial CD-NTase anti-phage defense enzymes [24]. Animal cGLRs form a single family of signaling proteins that share more distantly related homologies with bacterial CD-NTase anti-phage defense enzymes and animal OAS1-like proteins [18,24][18][24]. Furthermore, the interaction between STING and CDNs is universally conserved. However, some rare bacteriophages avoid potent innate immune responses by preventing cGAS/STING signaling through major capsid gene mutations in the CBASS [38,39,40][38][39][40]. Even bacteriophages escaping the bacterial cGLR or CBASS generate longer cabRNA that prevents the recognition and activation of CdnEO3.
STING-mediated CDN recognition also originated in bacteria as a defense mechanism against bacteriophages. Bacterial STING (b-STING) forms protein filaments to drive oligomerization of Toll/Interleukin 1 receptor (TIR) effector domains and rapid NAD+ cleavage [23]. STINGs and bacterial-like STING (bl-STINGs, present in microeukaryotes clustered between bacterial and metazoan sequences) share similarities in their domains [18]. Most new eukaryotic bl-STINGs have four N-terminal alpha helices as seen in human STING (h-STING). Furthermore, bacterial transmembrane domain -STINGs (TM-STINGS) have two N-terminal alpha helices and are more similar to bacterial Toll/Interleukin 1 receptor (TIR)-STING or TIR-STING, indicating eukaryotes and bacteria independently converged on a typical TM-STING domain architecture through domain shuffling [18]. Notably, TIR domains of oyster TIR-STING are related to animal TIRs and differ from the TIR domains of bacterial TIR-STINGs. Eukaryotic TIR-STINGs are rare, supporting the hypothesis that this protein originated from recent animal convergence or convergent domain shuffling [18,41][18][41]. Therefore, TM-STING and TIR-STING proteins have evolved due to two independent convergent evolution processes, and bacteria and eukaryotes have used similar proteins by reusing ancient protein domains.
The Drosophila STING (DmSTING or CG1667) is an ortholog to the vertebrate STING. DmSTING has evolutionarily conserved antibacterial (Listeria monocytogenes) and antiviral (positive-strand RNA Drosophila C virus and other DNA viruses) activity that involves the downstream activation of immune deficiency (IMD) and the Drosophila inhibitory kinase kinase β (dIKKβ) pathway and the subsequent cleavage of Relish (an NF-κB homolog) to generate antimicrobial peptides (AMPs) without the involvement of its cGAS ortholog CG7194. CG7194 lacks both the Zn2+-ribbon domain and a positively charged N terminus which are critical for DNA binding [42,43,44,45,46][42][43][44][45][46]. Furthermore, DmSTING transferred to mammalian cells induces an NF-κB-dependent immune response. Hence, Drosophila cGLRs, especially cGLR1, sense cytosolic dsRNA to generate the 3′2′-cGAMP that DmSTING recognizes to initiate the downstream innate immune response through activating Relish [47,48][47][48].
Drosophila cGLR2 generates 2′3′-cGAMP and 3′2′-cGAMP through an unknown target to generate a protective antimicrobial immune response [48]. The loss of DmSTING in Parkin RBR E3 ubiquitin-protein ligase KO (parkin−/−) flies rescues thorax muscle defects, climbing ability, and disrupted mitochondrial morphology of their flight muscles, indicating a critical noncanonical role of DmSTING in cellular and organismal responses to mitochondria stress [49]. Flies lacking parkin and DmSTING exhibit PTEN-induced kinase 1 (PINK1) activation to suppress cell death. Parkin, PINK1, and mitochondrial fidelity play critical roles in the pathogenesis of Parkinson’s disease (PD) in humans [50,51,52][50][51][52]; therefore, it would be interesting to see if a similar h-STING-Parkin-PINK-1 axis exists in human patients with PD. Thus, the cGAS/STING signaling-mediated immune defense mechanism is evolutionarily very ancient, with its origin dating back to prokaryotes/bacteria fighting against invading viruses. Eukaryotes, including humans, have acquired it via convergent domain shuffling and multi-HGT to guard against viruses and self-derived cytosolic DNAs.

Figure 1. Schematic representation of mammalian cGLR (cGAS/STING signaling) activation and downstream effects. cGAS recognizes cytosolic dsDNA, which can be foreign (pathogen-derived) or self- or adjacent-cell-generated (Mt-DNA and micronuclei generated due to genotoxic or oxidative stress) as a PAMP/DAMP. The enzymatic activity of cGAS cleaves cytosolic dsDNA into CDNs called cGAMP. STING recognizes cGAMP, which induces its transfer to ERGIC and the Golgi apparatus. For example, in the non-activated stage, it remains in the ER via binding to STIM-1 to retain its position in the ER membranes. The STING translocation to the Golgi apparatus phosphorylates (denoted by “P” in figure),TBK1 which induces IRF3 phosphorylation and induction of IRF3-dependent type 1 IFN genes. Active STING also phosphorylates TRAF6, which induces downstream signaling molecules (NIK and IκB) to initiate the transcription of NF-κB-dependent pro-inflammatory genes. Cells such as immune cells also express cGAMP importers, which import cGAMP generated by distant cells to initiate STING signaling without the involvement of cGAS itself. Notably, cGAS located in the nucleus is unable to recognize and bind to nuclear DNA to initiate the pro-inflammatory immune response. Kindly see the text for details.
References
- Li, D.; Wu, M. Pattern recognition receptors in health and diseases. Signal Transduct. Target. Ther. 2021, 6, 291.
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384.
- Gong, T.; Liu, L.; Jiang, W.; Zhou, R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol. 2020, 20, 95–112.
- Kumar, V. Toll-Like Receptors in Adaptive Immunity. Handb. Exp. Pharmacol. 2022, 276, 95–131.
- Kumar, V.; Barrett, J.E. Toll-Like Receptors (TLRs) in Health and Disease: An Overview. Handb. Exp. Pharmacol. 2022, 276, 1–21.
- Kumar, V. Toll-like receptors in the pathogenesis of neuroinflammation. J. Neuroimmunol. 2019, 332, 16–30.
- Kumar, V. Toll-like receptors in immunity and inflammatory diseases: Past, present, and future. Int. Immunopharmacol. 2018, 59, 391–412.
- Sharma, N.; Saxena, S.; Agrawal, I.; Singh, S.; Srinivasan, V.; Arvind, S.; Epari, S.; Paul, S.; Jha, S. Differential Expression Profile of NLRs and AIM2 in Glioma and Implications for NLRP12 in Glioblastoma. Sci. Rep. 2019, 9, 8480.
- Chou, W.-C.; Jha, S.; Linhoff, M.W.; Ting, J.P.Y. The NLR gene family: From discovery to present day. Nat. Rev. Immunol. 2023, 23, 635–654.
- Duxbury, Z.; Wu, C.-h.; Ding, P. A Comparative Overview of the Intracellular Guardians of Plants and Animals: NLRs in Innate Immunity and Beyond. Annu. Rev. Plant Biol. 2021, 72, 155–184.
- Van Gorp, H.; Kuchmiy, A.; Van Hauwermeiren, F.; Lamkanfi, M. NOD-like receptors interfacing the immune and reproductive systems. FEBS J. 2014, 281, 4568–4582.
- Brown, G.D.; Willment, J.A.; Whitehead, L. C-type lectins in immunity and homeostasis. Nat. Rev. Immunol. 2018, 18, 374–389.
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551.
- Loo, Y.M.; Gale, M., Jr. Immune signaling by RIG-I-like receptors. Immunity 2011, 34, 680–692.
- Davis, B.K.; Wen, H.; Ting, J.P. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 2011, 29, 707–735.
- Kumar, V. Inflammation research sails through the sea of immunology to reach immunometabolism. Int. Immunopharmacol. 2019, 73, 128–145.
- Canton, J.; Neculai, D.; Grinstein, S. Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 2013, 13, 621–634.
- Culbertson, E.M.; Levin, T.C. Eukaryotic CD-NTase, STING, and viperin proteins evolved via domain shuffling, horizontal transfer, and ancient inheritance from prokaryotes. PLoS Biol. 2023, 21, e3002436.
- Whiteley, A.T.; Eaglesham, J.B.; de Oliveira Mann, C.C.; Morehouse, B.R.; Lowey, B.; Nieminen, E.A.; Danilchanka, O.; King, D.S.; Lee, A.S.Y.; Mekalanos, J.J.; et al. Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 2019, 567, 194–199.
- Govande, A.A.; Duncan-Lowey, B.; Eaglesham, J.B.; Whiteley, A.T.; Kranzusch, P.J. Molecular basis of CD-NTase nucleotide selection in CBASS anti-phage defense. Cell Rep. 2021, 35, 109206.
- Davies, B.W.; Bogard, R.W.; Young, T.S.; Mekalanos, J.J. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 2012, 149, 358–370.
- Wu, X.; Wu, F.H.; Wang, X.; Wang, L.; Siedow, J.N.; Zhang, W.; Pei, Z.M. Molecular evolutionary and structural analysis of the cytosolic DNA sensor cGAS and STING. Nucleic Acids Res. 2014, 42, 8243–8257.
- Morehouse, B.R.; Govande, A.A.; Millman, A.; Keszei, A.F.A.; Lowey, B.; Ofir, G.; Shao, S.; Sorek, R.; Kranzusch, P.J. STING cyclic dinucleotide sensing originated in bacteria. Nature 2020, 586, 429–433.
- Li, Y.; Slavik, K.M.; Toyoda, H.C.; Morehouse, B.R.; de Oliveira Mann, C.C.; Elek, A.; Levy, S.; Wang, Z.; Mears, K.S.; Liu, J.; et al. cGLRs are a diverse family of pattern recognition receptors in innate immunity. Cell 2023, 186, 3261–3276.e20.
- Millman, A.; Melamed, S.; Amitai, G.; Sorek, R. Diversity and classification of cyclic-oligonucleotide-based anti-phage signalling systems. Nat. Microbiol. 2020, 5, 1608–1615.
- Burdette, D.L.; Monroe, K.M.; Sotelo-Troha, K.; Iwig, J.S.; Eckert, B.; Hyodo, M.; Hayakawa, Y.; Vance, R.E. STING is a direct innate immune sensor of cyclic di-GMP. Nature 2011, 478, 515–518.
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013, 339, 786–791.
- Banh, D.V.; Roberts, C.G.; Morales-Amador, A.; Berryhill, B.A.; Chaudhry, W.; Levin, B.R.; Brady, S.F.; Marraffini, L.A. Bacterial cGAS senses a viral RNA to initiate immunity. Nature 2023, 623, 1001–1008.
- Cohen, D.; Melamed, S.; Millman, A.; Shulman, G.; Oppenheimer-Shaanan, Y.; Kacen, A.; Doron, S.; Amitai, G.; Sorek, R. Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature 2019, 574, 691–695.
- Kumar, V. The Trinity of cGAS, TLR9, and ALRs Guardians of the Cellular Galaxy against Host-Derived Self-DNA. Front. Immunol. 2021, 11, 624597.
- Chow, K.L.; Hall, D.H.; Emmons, S.W. The mab-21 gene of Caenorhabditis elegans encodes a novel protein required for choice of alternate cell fates. Development 1995, 121, 3615–3626.
- Li, S.; Yang, F.; Wang, F.; Lv, X.; Li, F. An invertebrate gene encoding a Mab21-containing protein involves in antiviral response through regulating the STING pathway. Dev. Comp. Immunol. 2021, 121, 104101.
- Hornung, V.; Hartmann, R.; Ablasser, A.; Hopfner, K.-P. OAS proteins and cGAS: Unifying concepts in sensing and responding to cytosolic nucleic acids. Nat. Rev. Immunol. 2014, 14, 521–528.
- Wang, Y.; Holleufer, A.; Gad, H.H.; Hartmann, R. Length dependent activation of OAS proteins by dsRNA. Cytokine 2020, 126, 154867.
- Kristiansen, H.; Gad, H.H.; Eskildsen-Larsen, S.; Despres, P.; Hartmann, R. The oligoadenylate synthetase family: An ancient protein family with multiple antiviral activities. J. Interferon Cytokine Res. 2011, 31, 41–47.
- Zhang, X.; Wu, J.; Du, F.; Xu, H.; Sun, L.; Chen, Z.; Brautigam, C.A.; Zhang, X.; Chen, Z.J. The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop. Cell Rep. 2014, 6, 421–430.
- Li, X.; Shu, C.; Yi, G.; Chaton, C.T.; Shelton, C.L.; Diao, J.; Zuo, X.; Kao, C.C.; Herr, A.B.; Li, P. Cyclic GMP-AMP Synthase Is Activated by Double-Stranded DNA-Induced Oligomerization. Immunity 2013, 39, 1019–1031.
- Huiting, E.; Cao, X.; Ren, J.; Athukoralage, J.S.; Luo, Z.; Silas, S.; An, N.; Carion, H.; Zhou, Y.; Fraser, J.S.; et al. Bacteriophages inhibit and evade cGAS-like immune function in bacteria. Cell 2023, 186, 864–876.e21.
- Eaglesham, J.B.; Kranzusch, P.J. Conserved strategies for pathogen evasion of cGAS-STING immunity. Curr. Opin. Immunol. 2020, 66, 27–34.
- Ahn, J.; Barber, G.N. STING signaling and host defense against microbial infection. Exp. Mol. Med. 2019, 51, 1–10.
- Burroughs, A.M.; Aravind, L. Identification of Uncharacterized Components of Prokaryotic Immune Systems and Their Diverse Eukaryotic Reformulations. J. Bacteriol. 2020, 202, e00365-20.
- Martin, M.; Hiroyasu, A.; Guzman, R.M.; Roberts, S.A.; Goodman, A.G. Analysis of Drosophila STING Reveals an Evolutionarily Conserved Antimicrobial Function. Cell Rep. 2018, 23, 3537–3550.e6.
- Zakovic, S.; Levashina, E.A. Insects Go on a STING Operation to Tackle Intracellular Invaders. Immunity 2018, 49, 195–197.
- Goto, A.; Okado, K.; Martins, N.; Cai, H.; Barbier, V.; Lamiable, O.; Troxler, L.; Santiago, E.; Kuhn, L.; Paik, D.; et al. The Kinase IKKβ Regulates a STING- and NF-κB-Dependent Antiviral Response Pathway in Drosophila. Immunity 2018, 49, 225–234.e4.
- Dostert, C.; Jouanguy, E.; Irving, P.; Troxler, L.; Galiana-Arnoux, D.; Hetru, C.; Hoffmann, J.A.; Imler, J.-L. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat. Immunol. 2005, 6, 946–953.
- Lamiable, O.; Kellenberger, C.; Kemp, C.; Troxler, L.; Pelte, N.; Boutros, M.; Marques, J.T.; Daeffler, L.; Hoffmann, J.A.; Roussel, A.; et al. Cytokine Diedel and a viral homologue suppress the IMD pathway in Drosophila. Proc. Natl. Acad. Sci. USA 2016, 113, 698–703.
- Slavik, K.M.; Morehouse, B.R.; Ragucci, A.E.; Zhou, W.; Ai, X.; Chen, Y.; Li, L.; Wei, Z.; Bähre, H.; König, M.; et al. cGAS-like receptors sense RNA and control 3′2′-cGAMP signalling in Drosophila. Nature 2021, 597, 109–113.
- Holleufer, A.; Winther, K.G.; Gad, H.H.; Ai, X.; Chen, Y.; Li, L.; Wei, Z.; Deng, H.; Liu, J.; Frederiksen, N.A.; et al. Two cGAS-like receptors induce antiviral immunity in Drosophila. Nature 2021, 597, 114–118.
- Moehlman, A.T.; Kanfer, G.; Youle, R.J. Loss of STING in parkin mutant flies suppresses muscle defects and mitochondria damage. PLoS Genet. 2023, 19, e1010828.
- O’Callaghan, B.; Hardy, J.; Plun-Favreau, H. PINK1: From Parkinson’s disease to mitophagy and back again. PLOS Biol. 2023, 21, e3002196.
- Pickrell, A.M.; Youle, R.J. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 2015, 85, 257–273.
- Quinn, P.M.J.; Moreira, P.I.; Ambrósio, A.F.; Alves, C.H. PINK1/PARKIN signalling in neurodegeneration and neuroinflammation. Acta Neuropathol. Commun. 2020, 8, 189.
More
