2. Contamination of Toilet Bowl, Seat and Rim
Several studies have shown a correlation between hospital-acquired infections caused by MDR and contamination of different parts of the toilet. In most cases, such a relation was the cause of outbreak persistence or recurrence and represented a serious concern for patient clinical outcomes.
A study by Smismans and colleagues reported on the first detection of carbapenemase-producing
C. freundii in toilet bowls and the subsequent hospital-associated transmission to patients in one hospital ward
[19]. By sampling of high-touch surfaces, medical tools and toilet water, the
resea
rcheuthors found the latter to be positive for
C. freundii carrying the
blaOXA-48 gene. Admission in the room where OXA-48-producing
C. freundii was found in the toilet water was a risk factor for infection. Toilet cleaning with biguanide/quaternary ammonium for 15 min, followed by disinfection with 2500 parts per million (ppm) peracetic acid for 30 min was effective in eradicating the strain from the toilet and terminating the outbreak
[19].
Similarly, environmental investigations at a haematological ward of a French university hospital identified seven of 74 toilet rims positive for
blaOXA-48-positive CPE, including
C. freundii,
Enterobacter sakazakii and
E. coli [20]. Transmission of clonally related OXA-48-producing
C. freundii from the hospital environment to patients was demonstrated by whole genome comparisons. The majority of the clinical and environmental isolates of OXA-48-producing
C. freundii belonged to ST22 and were considered highly related by genome sequencing analysis (<50 SNPs between different strains)
[20]. It is interesting to note that, despite the concomitant circulation of NDM-type-producing Enterobacterales in the same ward, only
blaOXA-48-positive CPE were found in toilets, while strains harbouring
blaNDM-type genes were obtained from human samples only, demonstrating a higher environmental persistence ability of
blaOXA-48-positive organisms as compared to other CPE.
Among MDR bacteria colonizing the gut of inpatients, vancomycin-resistant Enterococci (VRE) should be mentioned, especially due to their increasing isolation in hospital settings
[21]. In a large study carried out in a non-outbreak setting, the contamination of several environmental sites was found. In particular, 13% of sampled toilet bowl seats were positive for at least an MDR organism (CPE, VRE and/or ESBL-producing Enterobacterales). The rate of environmental contamination was higher for patients with VRE (VanA-producing Enterococci) compared to other MDR organisms. Environmental samples from rooms housing VRE-colonized patients were positive in the 42% of cases
[22]. Vancomycin-resistant
Enterococcus faecium (VREfm) may also shed into the hospital environment, where it may persist despite standard cleaning. Indeed, Noble et al. reported that a hospital toilet represents a transmission vector for VREfm, since it facilitates the patient-to-patient transmission of
E. faecium [23]. For this reason, being admitted to a room previously occupied by a VREfm-positive patient is a risk factor for acquiring VREfm. Therefore, preventing the
E. faecium acquisition requires an understanding of reservoirs and transmission routes.
The role of the environment as a source of infection is even more relevant for spore-forming bacteria, such as
Clostridioides difficile. It has been estimated that a patient with
Clostridioides difficile infection (CDI) can excrete between 10
4 and 10
7 of
C. difficile per gram of faeces
[24]. Indeed, in a study performed by Reigadas et al., the toilet was the second most contaminated site with toxigenic
C. difficile [25]. The different ribotypes recovered corresponded mostly to frequent ribotypes in Spain
[26].
3. Contamination of Toilet Plumbing Systems
Some studies reported circulation and transmission of CPE originating form hospital plumbing installations
[27][28][29][30][31][32][33][34][27,28,29,30,31,32,33,34].
A high level of sinks siphons contamination by OXA-48-producing
K. pneumoniae and
E. coli was found in a traumatology and oncology ward, with patients admitted at rooms contaminated by OXA-48-producing CPE more frequently colonized
[29].
A similar scenario involving other CPE was observed in a burn unit in Belgium, where an outbreak of infections by OXA-48-producing
K. pneumoniae occurred
[30]. The concomitant presence of these bacteria in the toilet water of multiple rooms was observed, with the single origin of the outbreak confirmed by whole-genome multi-locus sequence typing (wgMLST) analysis. In fact, all outbreak isolates belonged to the same ST (i.e., ST15) and showed isogenicity (<15 allele differences)
[30]. Patients were not transferred in other rooms and, conversely, several drainpipe obstructions with consequent water reflux to the different toilets were reported, and thus, it was likely that OXA-48-producing
K. pneumoniae may have spread between different rooms through the common wastewater plumbing in a retrograde manner. The daily disinfection with bleach for two months was not sufficient to terminate the outbreaks, indicating that this method is insufficient to eradicate the strain in toilet water
Likewise, Hamerlinck et al. found toilet water, sink drains and shower drains mostly contaminated with OXA-48-producing clones (69.9%), followed by VIM (37%) and NDM-1 (1.4%).
C. freundii was the predominant species (52.1%), followed by
E. cloacae complex (41.1%),
K. pneumoniae (9.6%) and
K. oxytoca (6.8%)
[28]. Despite infection control measures and appropriate cleaning protocols, a long-term co-existence of five different OXA-48-producing
C. freundii clusters (ST22, ST170 and ST421, ST481 and ST146) was detected by a retrospective analysis using whole-genome sequencing (WGS) data
[28]. Such clusters involved both the patient as well as environmental isolates. Since patient-to-patient transmission was excluded due to long free intervals between stays, sanitary facilities may have played a role in the circulation of these clones.
A recent study in an intensive care unit (ICU) has also demonstrated that sinks traps located near toilets were more frequently contaminated by microorganisms carrying the
blaKPC gene (i.e., carbapenem-resistant
K. pneumoniae and
E. cloacae) than sink drains located away from toilets
[31]. This phenomenon could be multifactorial and to involve not only biofilm growth in communal pipes between toilets and sinks, but also the generation of contaminated droplets during flushing and/or the conduct of routine hand hygiene by patients or healthcare workers.
Other studies reported sinks as environmental reservoirs of CPE encoding for the
blaIMP-4 (i.e.,
E. cloacae complex ST24,
C. freundii ST8)
[32]. In a retrospective outbreak investigation carried out by WGS of stored CPE isolates from Australia, Marmor and colleagues revealed cases of
E. cloacae complex ST24 among both environmental samples and patients, while cases of
C. freundii ST8
blaIMP-4 only among patients
[32].
The healthcare wastewater drainage systems (i.e., drains, sink/shower siphons, drainage pipes), can be a reservoir for other pathogens, such as non-fermenting Gram-negative (NFGN) bacteria. Irregular designs of sinks and toilets, frequent blockages and leaks from waste pipes are factors that can contribute to the contamination
[35]. This is the case of several outbreaks caused by MDR
P. aeruginosa, where the hospital wastewater system was identified as a probable source of infection
[33][34][33,34]. Two outbreaks in England, one hospital-wide and the other one limited to a specialized medical unit, were caused by a different
P. aeruginosa genotype, both producers of VIM-2 carbapenemase
[33]. Aspelund and colleagues also described a prolonged nosocomial outbreak of VIM-producing
P. aeruginosa, where sink drains served as a long-term reservoir. In particular, the majority of strains isolated from sink showed the same antibiotic susceptibility phenotype and were identical or closely related (i.e., they showed the same or very similar pulsed-field gel electrophoresis (PFGE) band pattern) to clinical isolates
[34], demonstrating how the hydraulic environment could have played a role in establishing the outbreak.
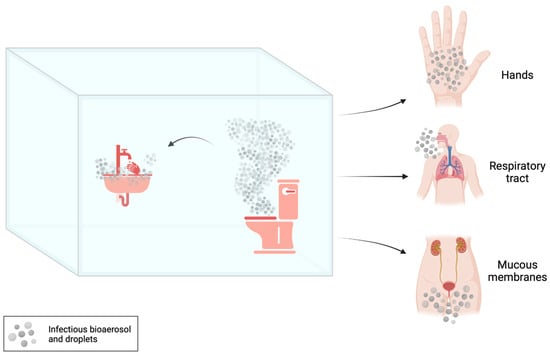
4. Contamination of Toilet Flushing-Generated Bioaerosol
In the context of shared toilets, bioaerosols can play an important role in the transmission of infections. When a toilet is flushed, a turbulent flow of water generates aerosols and droplets that can carry microorganisms and spores from faecal matter both into the air and the surrounding environment
[12][15][36][12,15,36].
In a pre/post-intervention design study, conducted in adult intensive care units (ICUs) of a university hospital, the installation of hopper covers (a “toilet-like” waste disposal system) and sink trap devices were effective in reducing the acquisitions of carbapenemase-producing Enterobacterales (CPE) through particles dispersion after toilet flushing
[37]. The
res
earchtudy further confirmed the role of hospital wastewater plumbing as a reservoir in nosocomial transmission of multispecies CPE (e.g.,
Serratia marcescens,
Enterobacter cloacae complex,
Citrobacter freundii and
Klebsiella oxytoca-producing KPC-type carbapenemases)
[37].
In another study, reproducing bacterial loads observed during symptomatic phase of
Clostridioides difficile infection (CDI), Best et al. found airborne spread and consequent environmental contamination of this pathogen through the aerosol produced following flushing in different types of toilets (with and without lid) typically used in hospital settings
[38]. The
resea
rcheuthors detected high levels of
C. difficile, especially in toilets without lids, after flushing the toilet. The surface contamination near the toilet has also been confirmed
[38]. Similarly, the air of bathrooms used by patients with CDI were sampled both before and after flushing by Wilson et al.
[39]. The
resea
rcheuthors found that pre- and post-flush samples were positive for
Enterococcus faecalis,
E. faecium and
C. difficile, with major bacterial concentrations observed in post-flush samples
[39].