Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Camila Xu and Version 1 by Jasjit Suri.
The ability of UNet-based deep learning models as shown before is very powerful in the imaging domain and can handle image noise, structure, scale, size, resolution, and further, the variability in the shapes.
- coronary artery disease
- intravascular ultrasound
- deep learning
1. Introduction
One of the world’s greatest contributors to mortality and morbidity is cardiovascular disease (CVD), which accounts for about 18 million deaths per year [1]. The primary two causes of CVD-related fatalities are coronary artery disease (CAD) and acute coronary syndrome (ACS) [2]. Generally speaking, CAD entails the shrinking of arteries as a result of the buildup of atherosclerotic plaque within their walls, resulting in coronary artery obstruction [3]. Aiming to enhance the diagnosis and treatment of heart disorders as well as lowering the fatality rate from CVD, significant advancements have been made in cardiovascular research and therapy in recent decades [4]. It is now possible to carry out a comprehensive qualitative and quantitative assessment of heart morphological structures as well as operations with the use of contemporary medical imaging techniques, including intravascular ultrasound (IVUS) [5,6,7[5][6][7][8][9],8,9], computed tomography (CT) [10], magnetic resonance imaging (MRI) [11[11][12][13],12,13], and ultrasound (US) [14[14][15],15], which assist identification, disease monitoring, surgical planning, and evaluation. An example of the coronary artery is shown in Figure 1a, while the IVUS acquisition device for the coronary vascular system is shown in Figure 1b.
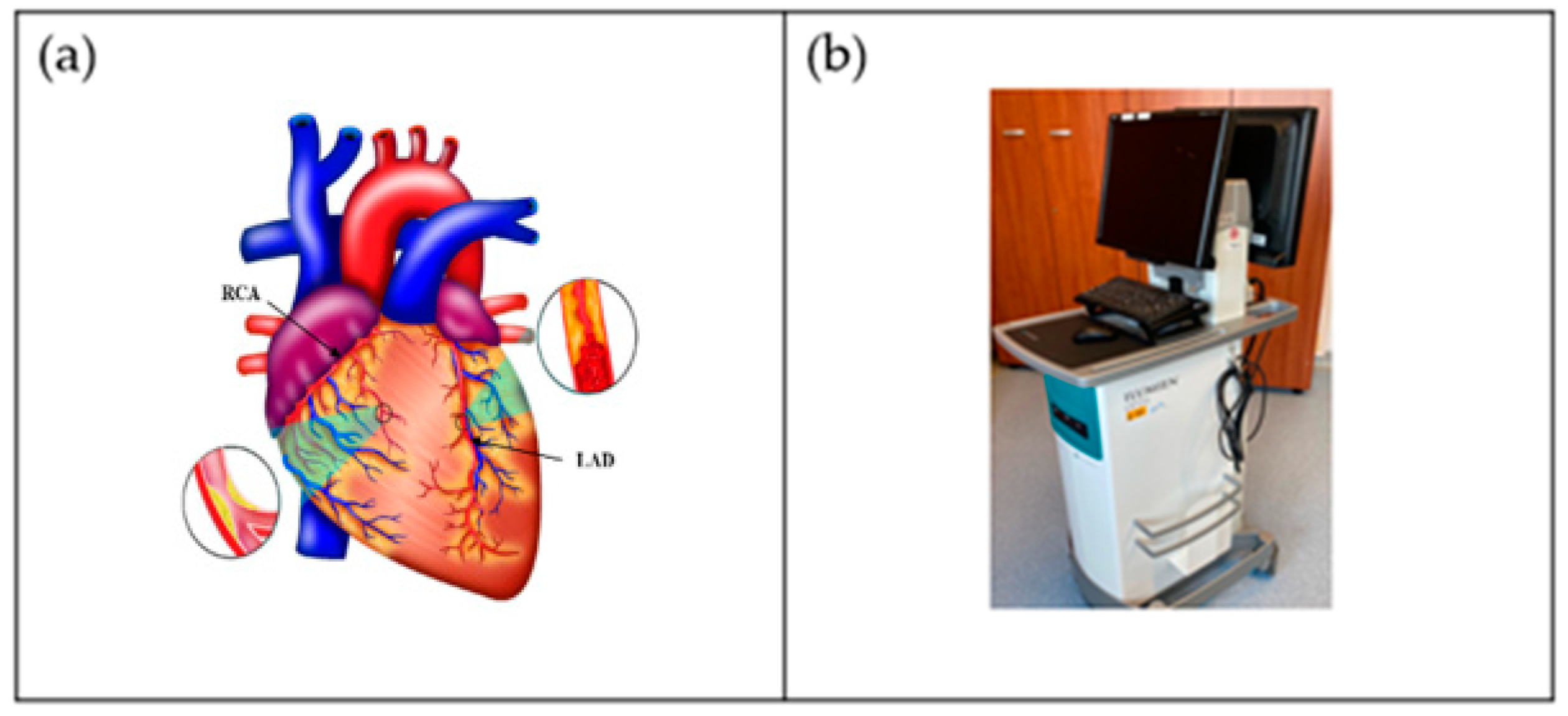
Figure 1. (a) Coronary arteries of the heart showing LAD (left anterior descending coronary artery) and RCA (right coronary artery) (Courtesy of AtheroPoint™, Roseville, CA, USA). (b) IVUS acquisition device (Courtesy of Dr. Alberto Boi and Luca Saba, University of Cagliari, Cagliari, Italy).
The diagnosis of CAD is frequently made by coronary CT angiography (CCTA), which enables non-invasive measurement of the arterial lumen’s diameter and plaque localization [16,17,18,19,20][16][17][18][19][20]. However, radiologists presently manually assess the location and severity of the plaque(s) leading to the stenosis in CCTA pictures, which, in addition to being costly and time-consuming, is also susceptible to mistake and inaccuracy [21]. In order to develop computerized and accurate coronary artery stenosis as well as a plaque identification method, it is crucial that coronary arteries in CCTA pictures must be automatically segmented. The following factors, however, make automatic coronary artery segmentation for CCTA pictures particularly complicated. To begin with, coronary circulation has a complicated pattern, with several arteries of different thicknesses [22]. For perfect segmentation, some of the branches are even too thin. Additionally, individual differences in the structure of the coronary artery tree may be relevant. Second, other vascular organs that seem like the coronary arteries adjacent to the heart can be mistaken for them because of their similar appearance [23]. Third, the coronary arteries only make up a tiny fraction of the entire heart’s cells, and the methods for segmentation must consider this imbalance [24]. Additionally, several variables, including heart rate, the data reconstruction method, the quantity of the injected contrast agent, and radiation exposure, affect the quality of the pictures obtained during CT angiography [25]. Coronary artery segmentation, therefore, is more challenging due to low-resolution image quality.
Figure 1b shows a popular imaging method for the assessment and control of CVD, intravascular ultrasound (IVUS) [5,26][5][26]. In conjunction with positional data, IVUS images are segmented into interior and exterior regions as lumen and media regions, respectively. Arteries’ representation in 3D heavily depends on arterial vessel walls for various purposes such as surgical planning. The arteries’ segmentation is helpful for plaque identification in clinical practices. IVUS-guided percutaneous coronary intervention (PCI) is a more advanced and superior technique in comparison to standard angiography-guided PCI, minimizing death risks in patients [6]. IVUS segmentation for lumen and vessel cross-sectional based on 3D vessel reconstruction is precise and quick for accurate and real-time segmentation during PCI [27]. However, IVUS segmentation requires recent, accurate, and faster techniques, typically at 30 Hz and 100 Hz frame rates. To record an IVUS sequence, a catheter-borne ultrasound transducer is inserted into the coronary artery and then returned via arteries at a speed of roughly 1 mm/s [5]. Raw radio frequency (RF) information from the probe is typically not used for analysis. However, amplified and filtered gray-scale B-mode Euclidean ultrasound pictures showing the coronary cross-section provide a typical output format for downstream evaluation (see Figure 2) The arrows in Figure 2 depict a typical example of five (1–5) frames with calcified plaques. Six patients’ IVUS videos’ worth of frames were collected, and they were placed in a 6 × 5 matrix. The symbol I (1,1)-I (6,5) is used to represent this.
IVUS segmentation is one of the most challenging tasks in medical images. It consists of lumen–intima (LI) and media–adventitia (MA) border detection. This challenge is due to the presence of the artifacts, namely shadows, bifurcation, and echogenic plaques, and the fact that public expert-labeled ground-truth databases only contain a small number of captures [28]. Even though artificial intelligence (AI) has shown promising signs toward higher accuracy and learning strategy, it has been observed that these AI-based black boxes lack clinical validation and the ability to perform well in clinical settings, and they are unable to explain the outcomes [29,30,31,32,33,34,35][29][30][31][32][33][34][35]. The clinical validation requires that the outcome from the AI system must have a behavior leading to correct coronary artery disease risk assessment. For example, should an AI system perform accurately on a test patient who has a high risk, then the syntax score of this patient should be high [36]. Other ways to show the clinical validation include by estimating the relationships or correlations between two quantities such as computed tomography (CT) coronary artery score vs. AI outcome of the risk [37]. Such consistent behavior needs to be exhibited by AI systems. Other than the clinical validation, there are attributes such as imbalanced classes in the datasets that can introduce AI bias [38]. Such causes can lead to bias in AI modules or system designs.
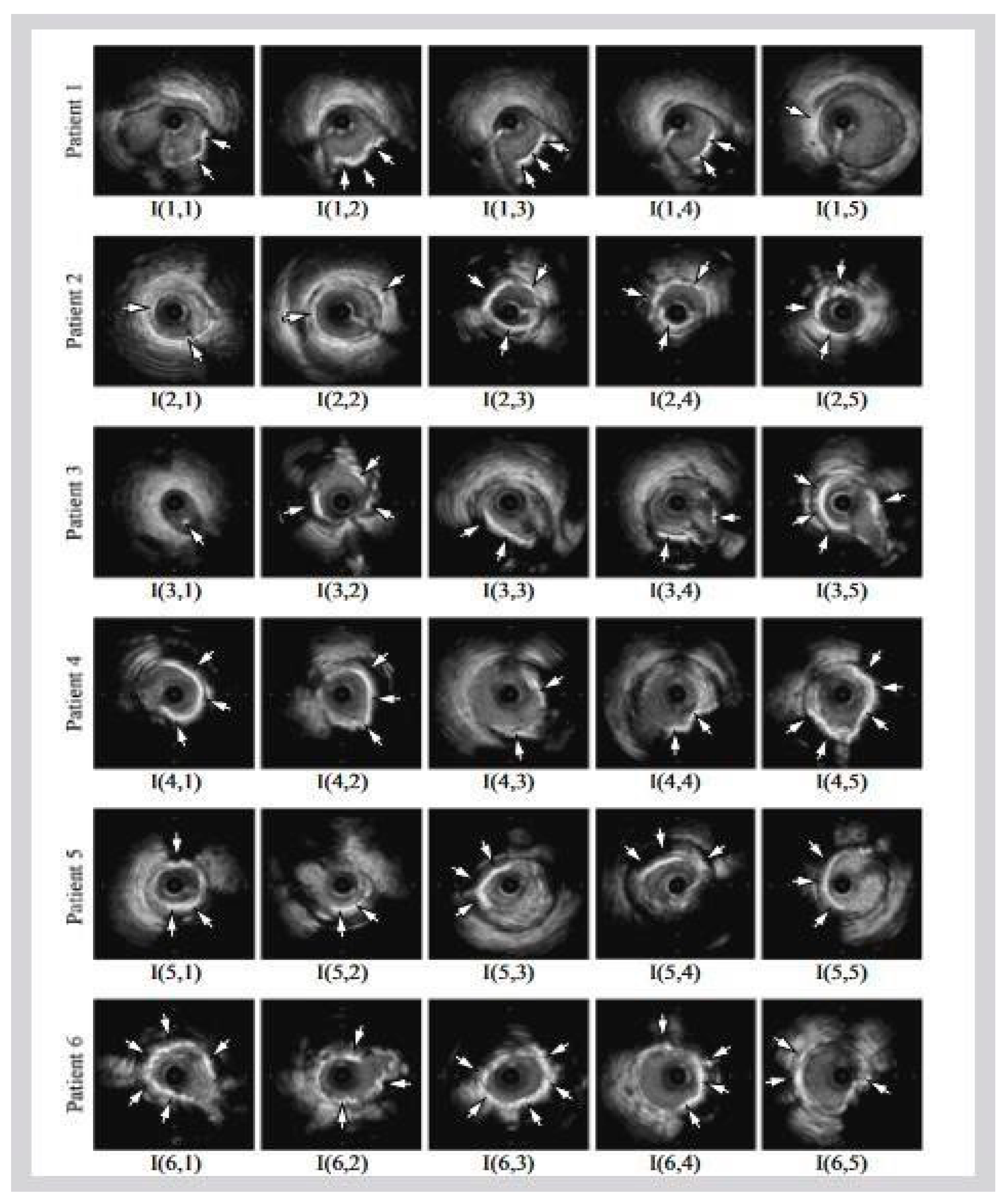
Figure 2. Six patients’ calcified plaques can be seen in sample frames I (1,1) through I (6,5) from the overall intravascular ultrasonography (IVUS) films [43][39].
2. Characteristics of UNet and Conventional DL Systems for CAD
2.1. A Special Note on Limitations of Conventional Models and Benefits of AI-Based Solutions
The conventional models adopted in image processing have existed for the last 50 years [130,131,132,133,134][40][41][42][43][44]. These methods were considered as generation I and II, where the methods were considered as local in nature and never used the cohort’s knowledge for the benefit for prediction on the test datasets. These methods had some inherent drawbacks, such as inability to provide an automated solution towards segmentation of the organs in complex medical images [130,135,136][40][45][46]. These methods were local in nature, and the noise would overwhelming and distract the computer vision algorithms [137][47]. Thus, the system was ad hoc in nature and could not be automated for every new incoming test image [138][48]. Due to these inherent challenges, the performance in these systems dropped considerately and affected the accuracy, sensitivity, specificity, Mathew coefficient, recall, precision, area under the curve, and p-value significance. Further, the statistical tests for evaluating the reliability and stability also did not perform well, which included the t-test, paired t-test, Bonferroni tests, Freedman test, Wilcoxon test, Poisson test, etc. [139,140,141][49][50][51]. The effect of such challenges lacked explainability and interpretations [41,142][52][53]. As a result, time and again, these computer vision methods started losing interest, and over time, inventions based on knowledge derived by the cohorts started to take shape.
With the invention of fundamental neural networks [143][54], these fundamental drawbacks started to disappear. The rapid rise of these methods has nearly dominated the field of image processing, which were then characterized into machine learning and deep learning approaches [144][55]. The most powerful paradigm was the addition of addition of intermediate layers between the input and output layers [145][56]. Deep learning solutions offer the following benefits over the conventional models: automated feature extraction, the power of the integration of knowledge from cohorts for better segmentation and classification solutions, the ability to adjust the depth of layers, the ability to parallelize these neural networks to improve the performance and optimize these deep layers, and the ability to reduce the noise present in the images using dropout layers.
2.2. A Special Note on Quality Control for AI Systems
The size of the cohort, the balancing of the class in the cohort, missing values in the cohort, scaled values of the risk factors, normalization of the factors if any, and augmentation of the raw datasets are all factors that are part of the quality control system during AI design. If the quality control is not conducted in a proper way, then the AI system may lack generalization. In other words, the training system will not be generalized. The cohort size plays a major role. If the cohort size is small, it can also cause overfitting. Thus, dropout layers help in improving the generalization. To further improve the generalization requires hyper-parameter tuning [145,150][56][57].
Table 1 tabulates the general characteristics of the DL system, described by using 26 attributes categorized into 5 clusters, namely demographic (rows A1–A3), architectural details of the deep learning model (rows A4–A10), performance evaluation (rows A11–A20), parameter optimization (rows A21–A25), and clinical evaluation (row A26). The cohort size used in different studies was very limited. The demographic factors considered by most of the studies were cohort size, smoking, and hypertension.
Table 1.
Characteristics of UNet and conventional systems for CAD.
| A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 | A9 | A10 | A11 | A12 | A13 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SN | Studies | TP | SM | HT | AU | L | EC | DC | SC | LF | PL | DSC | PV | JI |
| 1 | He et al. [44][58] | 2 | - | - | U | 5 | 3 | 3 | √ | √ | √ | √ | - | √ |
| 2 | Jin et al. [95][59] | 5 | - | - | C | - | - | - | - | - | - | - | √ | - |
| 3 | Ibtehaz et al. [45][60] | 1 | - | - | U | 5 | 5 | 5 | √- | √ | √ | - | - | √ |
| 4 | Balakrishna et al. [46][61] | 1 | - | - | U | 5 | 5 | 5 | √ | - | √ | √ | - | √ |
| 5 | Kim et al. [48][62] | 1 | - | - | U | 5 | 5 | 5 | √ | √ | √ | √ | - | √ |
| 6 | Li et al. [47][63] | 1 | - | - | U | 5 | 4 | 4 | √ | √ | √ | √ | - | - |
| 7 | Chen et al. [49][64] | 1 | - | - | U | 5 | 3 | 3 | √ | √ | √ | √ | - | - |
| 8 | Tong et al. [50][65] | 1 | - | - | U | 5 | 3 | 3 | √ | √ | √ | √ | - | - |
| 9 | Morris et al. [51][66] | 1 | - | - | U | 5 | 4 | 4 | √ | √ | √ | √ | - | - |
| 10 | Zhou et al. [52][67] | 1 | - | - | U | 5 | 5 | 5 | √ | √ | √ | √ | - | - |
| 11 | Milletari et al. [53][68] | 1 | - | - | U | 5 | 5 | 5 | √ | √ | - | √ | - | - |
| 12 | Szarski et al. [54][69] | 1 | - | - | U | 5 | 5 | 5 | √ | √ | √ | - | - | √ |
| 13 | Vercio et al. [65][70] | 0 | - | - | C | - | - | - | - | - | - | - | - | √ |
| 14 | Yang et al. [55][71] | 1 | - | - | U | 5 | 4 | 4 | √ | -√ | √ | - | - | √ |
| 15 | Shen et al. [56][72] | 5 | - | - | U | 5 | 4 | 4 | √ | √ | - | √ | - | √ |
| 16 | Javorszky et al. [57][73] | 5 | - | - | U | 5 | 4 | 4 | √ | √ | √ | - | √ | - |
| 17 | Momin et al. [58][74] | 5 | - | - | U | 5 | 3 | 3 | - | - | √ | √ | √ | - |
| 18 | Guo et al. [99][75] | 5 | √ | √ | U | 5 | 4 | 4 | √ | √ | √ | √ | √ | - |
| 19 | Huang et al. [67][76] | 5 | - | - | U | 5 | 5 | 5 | √ | √ | √ | √ | - | √ |
| 20 | Jun et al. [100][77] | 0 | - | - | U | 5 | 5 | 5 | - | √ | √ | √ | - | - |
| 21 | Shi et al. [64][78] | 0 | - | - | U | 5 | 5 | 5 | √ | √ | √ | - | - | - |
| 22 | Thuy et al. [68][79] | 0 | - | - | U | 5 | 4 | 4 | √ | √ | √ | √ | - | - |
| 23 | Hwang et al. [69][80] | 0 | - | - | U | 5 | 5 | 5 | - | - | - | - | - | - |
| 24 | Cheung et al. [60][81] | 5 | - | - | U | 5 | 5 | 5 | √ | - | √ | √ | - | - |
| 25 | Dong et al. [61][82] | 3 | - | - | U | 5 | 8 | 8 | √ | √ | - | - | - | √ |
| 26 | Pan et al. [70][83] | 0 | - | - | U | 5 | 4 | 4 | √ | √ | √ | √ | - | √ |
| 27 | Song et al. [24] | 5 | - | - | U | 5 | 4 | 4 | √ | √ | √ | √ | - | - |
| 28 | Shinohara et al. [62][84] | 3 | - | - | U | 5 | 5 | 5 | √ | - | √ | √ | √ | √ |
| 29 | Yang et al. [55][71] | 0 | - | - | U | 5 | 5 | 5 | √ | √ | √ | - | - | √ |
| 30 | Xia et al. [71][85] | 0 | - | - | U | 5 | 5 | 5 | √ | √ | √ | - | - | √ |
| 31 | Azad et al. [72][86] | 4 | - | - | U | 5 | 4 | 4 | √ | - | √ | - | - | - |
| 32 | Ronneberger et al. [73][87] | 0 | - | - | U | 5 | 4 | 4 | √ | √ | √ | - | - | - |
| 33 | Bajaj et al. [74][88] | 1 | - | √ | C | - | - | - | - | √ | - | - | √ | - |
| 34 | Cho et al. [75][89] | 5 | - | - | C | - | - | - | - | √ | - | - | √ | - |
| 35 | Araki et al. [26] | 2 | - | - | C | - | - | - | - | - | - | √ | √ | √ |
| 36 | Bajaj et al. [76][90] | 2 | - | - | C | - | - | - | - | √ | - | - | - | - |
| 37 | Fedewa et al. [77][91] | 0 | - | - | C | - | - | - | - | - | - | - | - | - |
| 38 | Masuda et al. [78][92] | 5 | - | - | C | - | - | - | - | √ | - | - | - | - |
| 39 | Min et al. [79][93] | 1 | - | √ | C | - | - | - | - | √ | - | - | √ | - |
| 40 | Nishi et al. [80][94] | 2 | - | - | C | - | - | - | - | √ | √ | √ | √ | - |
| 41 | Shin et al. [111][95] | 2 | - | - | C | - | - | - | - | - | - | - | √ | - |
| 42 | Olender et al. [81][96] | 1 | - | - | C | - | - | - | - | √ | - | - | √ | - |
| 43 | Zhao et al. [82][97] | 2 | - | - | C | - | - | - | - | - | - | - | √ | - |
| 44 | Bargsten et al. [83][98] | 1 | - | - | C | - | - | - | - | √ | - | √ | - | - |
| 45 | Samuel et al. [96][99] | 3 | - | √ | C | - | - | - | - | √ | √ | - | - | - |
| 46 | Faraji et al. [28] | 1 | - | - | C | - | - | - | - | - | - | - | - | √ |
| 47 | Tayel et al. [84][100] | 1 | - | - | C | - | - | - | - | - | - | - | - | - |
| 48 | Cui et al. [85][101] | 1 | - | - | C | - | - | - | - | - | - | - | - | √ |
| 49 | Harms et al. [86][102] | 5 | - | - | C | - | - | - | - | - | - | √ | √ | - |
| 50 | Mishra et al. [87][103] | 0 | - | - | C | - | - | - | - | √ | √ | √ | - | - |
| 51 | Lin et al. [97][104] | 5 | - | - | C | - | - | - | - | - | - | √ | √ | - |
| 52 | Du et al. [98][105] | 0 | - | - | C | - | - | - | - | √ | - | √ | - | √ |
| 53 | Hwang et al. [69][80] | 2 | - | - | C | - | - | - | - | - | - | - | - | - |
| 54 | Jodas et al. [88][106] | 0 | - | - | C | - | - | - | - | - | - | √ | - | √ |
| 55 | Eslamizadeh et al. [89][107] | 0 | - | - | C | - | - | - | - | - | - | - | - | - |
| 56 | Sofian et al. [90][108] | 1 | - | - | C | - | - | - | - | - | - | - | - | √ |
| 57 | Cao et al. [91][109] | 3 | - | - | C | - | - | - | - | - | - | √ | √ | √ |
| 58 | Taki et al. [92][110] | 1 | - | - | C | - | - | - | - | - | - | - | - | - |
| 59 | Unal et al. [93][111] | 0 | - | - | C | - | - | - | - | - | - | - | - | - |
| 60 | Zhu et al. [94][112] | 0 | - | - | C | - | - | - | - | - | - | - | - | - |
| A14 | A15 | A16 | A17 | A18 | A19 | A20 | A21 | A22 | A23 | A24 | A25 | A26 | ||
| SN | Studies | HD | Val | LR | BS | EPO | OPT | DA | Acc. | Pres. | RS | SN | SP | CE |
| 1 | He et al. [44][58] | √ | - | - | - | - | - | - | - | - | - | √ | √ | S |
| 2 | Jin et al. [95][59] | - | - | - | - | - | - | - | √ | √ | √ | √ | √ | M |
| 3 | Ibtehaz et al. [45][60] | - | - | - | - | √ | - | √ | √ | - | - | - | - | M |
| 4 | Balakrishna et al. [46][61] | - | - | √ | √ | √ | - | √ | √ | - | - | - | - | S |
| 5 | Kim et al. [48][62] | √ | √ | √ | √ | √ | - | - | - | - | - | - | - | S |
| 6 | Li et al. [47][63] | - | - | √ | √ | √ | √ | - | - | √ | - | √ | √ | S |
| 7 | Chen et al. [49][64] | - | - | - | - | - | - | √ | - | - | - | - | - | S |
| 8 | Tong et al. [50][65] | - | - | - | √ | √ | - | √ | - | - | - | - | - | S |
| 9 | Morris et al. [51][66] | - | - | √ | √ | √ | - | √ | - | - | - | - | - | S |
| 10 | Zhou et al. [52][67] | - | - | - | - | - | - | - | - | - | - | √ | √ | S |
| 11 | Milletari et al. [53][68] | √ | - | √ | √ | √ | - | √ | - | - | - | - | - | S |
| 12 | Szarski et al. [54][69] | √ | - | √ | √ | √ | √ | √ | - | - | - | - | - | S |
| 13 | Vercio et al. [65][70] | √ | - | - | - | - | - | - | - | - | - | - | - | S |
| 14 | Yang et al. [55][71] | √ | - | √ | √ | √ | √ | √ | - | - | - | - | - | S |
| 15 | Shen et al. [56][72] | - | - | √ | √ | √ | √ | - | - | - | - | - | - | S |
| 16 | Javorszky et al. [57][73] | - | - | - | - | - | √ | - | - | - | - | - | - | S |
| 17 | Momin et al. [58][74] | √ | - | √ | √ | √ | √ | √ | - | - | - | - | - | M |
| 18 | Guo et al. [99][75] | √ | √ | - | - | - | - | √ | - | - | - | - | - | S |
| 19 | Huang et al. [67][76] | - | - | √ | √ | √ | √ | √ | - | - | - | - | √ | S |
| 20 | Jun et al. [100][77] | - | - | √ | √ | √ | √ | √ | - | - | - | - | - | S |
| 21 | Shi et al. [64][78] | - | - | √ | - | √ | - | - | - | - | - | - | - | S |
| 22 | Thuy et al. [68][79] | - | - | √ | - | - | - | √ | - | - | - | - | - | S |
| 23 | Hwang et al. [69][80] | - | - | √ | √ | √ | - | - | - | - | - | - | - | S |
| 24 | Cheung et al. [60][81] | - | - | √ | - | √ | √ | - | - | - | - | - | - | S |
| 25 | Dong et al. [61][82] | - | - | √ | √ | - | √ | √ | - | - | - | - | - | S |
| 26 | Pan et al. [70][83] | - | - | - | - | - | - | - | - | - | - | - | - | S |
| 27 | Song et al. [24] | - | - | √ | √ | √ | √ | - | - | √ | √ | - | - | S |
| 28 | Shinohara et al. [62][84] | - | - | √ | √ | - | √ | √ | √ | √ | √ | - | - | S |
| 29 | Yang et al. [55][71] | √ | √ | √ | √ | √ | √ | √ | - | - | - | - | - | S |
| 30 | Xia et al. [71][85] | - | - | √ | √ | √ | √ | √ | - | - | - | - | - | S |
| 31 | Azad et al. [72][86] | - | - | - | - | √ | - | - | √ | - | - | √ | √ | S |
| 32 | Ronneberger et al. [73][87] | - | - | - | √ | - | - | √ | - | - | - | - | - | S |
| 33 | Bajaj et al. [74][88] | - | - | √ | √ | √ | - | - | √ | √ | - | - | - | S |
| 34 | Cho et al. [75][89] | - | √ | √ | - | √ | √ | √ | √ | - | - | √ | √ | S |
| 35 | Araki et al. [26] | - | - | - | - | - | - | - | - | - | √ | - | - | S |
| 36 | Bajaj et al. [76][90] | - | - | √ | √ | √ | - | √ | - | - | - | - | - | S |
| 37 | Fedewa et al. [77][91] | - | - | - | - | - | - | - | - | - | - | - | - | S |
| 38 | Masuda et al. [78][92] | - | - | √ | - | - | - | √ | √ | - | - | - | - | S |
| 39 | Min et al. [79][93] | - | √ | √ | - | - | - | √ | √ | √ | - | √ | √ | S |
| 40 | Nishi et al. [80][94] | - | - | √ | √ | √ | √ | - | - | - | - | - | - | S |
| 41 | Shin et al. [111][95] | - | - | - | - | - | - | - | - | - | - | - | - | S |
| 42 | Olender et al. [81][96] | - | - | √ | √ | √ | - | - | √ | √ | - | - | - | S |
| 43 | Zhao et al. [82][97] | - | √ | - | - | - | - | √ | √ | √ | √ | - | - | S |
| 44 | Bargsten et al. [83][98] | - | √ | √ | √ | √ | √ | - | - | - | - | - | - | S |
| 45 | Samuel et al. [96][99] | - | - | √ | √ | √ | - | √ | √ | - | - | √ | √ | M |
| 46 | Faraji et al. [28] | √ | - | - | - | - | - | - | - | - | - | - | - | S |
| 47 | Tayel et al. [84][100] | - | - | - | - | - | - | - | √ | - | - | - | - | S |
| 48 | Cui et al. [85][101] | - | - | - | - | - | - | - | - | - | - | - | - | S |
| 49 | Harms et al. [86][102] | √ | √ | √ | √ | √ | √ | - | - | - | - | - | - | S |
| 50 | Mishra et al. [87][103] | - | √ | √ | √ | √ | - | √ | - | - | - | - | - | S |
| 51 | Lin et al. [97][104] | - | - | - | - | - | - | - | - | - | - | √ | √ | M |
| 52 | Du et al. [98][105] | √ | - | - | √ | √ | √ | √ | - | - | - | - | - | M |
| 53 | Hwang et al. [69][80] | - | √ | - | - | - | - | - | - | - | - | √ | √ | S |
| 54 | Jodas et al. [88][106] | √ | - | - | - | - | - | - | - | - | - | - | - | S |
| 55 | Eslamizadeh et al. [89][107] | - | - | - | - | - | - | - | √ | - | - | - | - | S |
| 56 | Sofian et al. [90][108] | √ | - | - | - | - | - | - | - | - | - | - | - | S |
| 57 | Cao et al. [91][109] | - | - | - | - | - | - | - | - | - | - | - | - | S |
| 58 | Taki et al. [92][110] | - | - | - | - | - | - | - | - | - | - | - | - | S |
| 59 | Unal et al. [93][111] | - | - | - | - | - | - | - | - | - | - | - | - | S |
| 60 | Zhu et al. [94][112] | √ | - | - | - | - | - | - | - | - | - | - | - | S |
TP, total patients; SM, smoking; HT, hypertension; AU, architecture used; L, layers; EC, encoder; DC, decoder; SC, skip connection; LF, loss function; Pool, pooling; DSC, Dice similarity coefficient; SN, sensitivity; SP, specificity; JI, Jaccard index; HD, Hausdorff distance; Acc, accuracy; PV, p-value; Pres, precision; RS, recall score; Val, validation; LR, learning rate; BS, batch size; Epo, epochs; OPT, optimization; DA, data augmentation; CE, clinical evaluation; U, UNet; C, conventional; S, single center; M, multicenter; √ implies that a particular attribute (column) was implemented in that study (row).
The architectural details included in the AI-based system describe whether the given architecture is a conventional architecture or UNet architecture. The performance evaluation parameters used were DSC, sensitivity, specificity, Jaccard index, Hausdorff distance, p-value, accuracy, precision, and recall score. The parameter optimization in the DL system included learning rate, batch size, epochs, optimization, and data augmentation. The clinical evaluation considered single-center or multi-center data.
Standardized data augmentation was conducted on these images [40,148,151,152][113][114][115][116]. Data augmentation plays a crucial role in improving the generalization of machine learning models, including those used for coronary artery wall segmentation in intravascular ultrasound (IVUS) scans. These techniques help increase the diversity of the training data, making the model more robust to variations in the input data. Some specific data augmentation techniques commonly used in coronary artery wall segmentation for IVUS scans are as follows: (1) rotation from −50 to 100, (2) random flipping, (3) rotation to 2700, and (4) skewing [151][115].
References
- Smith, S.C., Jr.; Collins, A.; Ferrari, R.; Holmes, D.R., Jr.; Logstrup, S.; McGhie, D.V.; Ralston, J.; Sacco, R.L.; Stam, H.; Taubert, K. Our time: A call to save preventable death from cardiovascular disease (heart disease and stroke). Circulation 2012, 126, 2769–2775.
- Chan, M.Y.; Du, X.; Eccleston, D.; Ma, C.; Mohanan, P.P.; Ogita, M.; Shyu, K.-G.; Yan, B.P.; Jeong, Y.-H. Acute coronary syndrome in the Asia-Pacific region. Int. J. Cardiol. 2016, 202, 861–869.
- Suri, J.S.; Kathuria, C.; Molinari, F. (Eds.) Atherosclerosis Disease Management; Springer Science & Business Media: Berlin, Germany, 2010.
- Kandaswamy, E.; Zuo, L. Recent advances in treatment of coronary artery disease: Role of science and technology. Int. J. Mol. Sci. 2018, 19, 424.
- Katouzian, A.; Angelini, E.D.; Carlier, S.G.; Suri, J.S.; Navab, N.; Laine, A.F. A state-of-the-art review on segmentation algorithms in intravascular ultrasound (IVUS) images. IEEE Trans. Inf. Technol. Biomed. 2012, 16, 823–834.
- Darmoch, F.; Alraies, M.C.; Al-Khadra, Y.; Moussa Pacha, H.; Pinto, D.S.; Osborn, E.A. Intravascular ultrasound imaging–guided versus coronary angiography–guided percutaneous coronary intervention: A systematic review and meta-analysis. J. Am. Heart Assoc. 2020, 9, e013678.
- Escolar, E.; Weigold, G.; Fuisz, A.; Weissman, N.J. New imaging techniques for diagnosing coronary artery disease. CMAJ 2006, 174, 487–495.
- Banchhor, S.K.; Londhe, N.D.; Araki, T.; Saba, L.; Radeva, P.; Laird, J.R.; Suri, J.S. Well-balanced system for coronary calcium detection and volume measurement in a low resolution intravascular ultrasound videos. Comput. Biol. Med. 2017, 84, 168–181.
- Boi, A.; Jamthikar, A.D.; Saba, L.; Gupta, D.; Sharma, A.; Loi, B.; Laird, J.R.; Khanna, N.N.; Suri, J.S. A survey on coronary atherosclerotic plaque tissue characterization in intravascular optical coherence tomography. Curr. Atheroscler. Rep. 2018, 20, 1–17.
- Saba, L.; Suri, J.S. Multi-Detector CT Imaging: Principles, Head, Neck, and Vascular Systems; CRC Press: Boca Raton, FL, USA, 2013; Volume 1.
- Saba, L.; Agarwal, N.; Cau, R.; Gerosa, C.; Sanfilippo, R.; Porcu, M.; Montisci, R.; Cerrone, G.; Qi, Y.; Balestrieri, A. Review of imaging biomarkers for the vulnerable carotid plaque. JVS Vasc. Sci. 2021, 2, 149–158.
- Caredda, G.; Bassareo, P.P.; Cherchi, M.V.; Pontone, G.; Suri, J.S.; Saba, L. Anderson-fabry disease: Role of traditional and new cardiac MRI techniques. Br. J. Radiol. 2021, 94, 20210020.
- Cau, R.; Cherchi, V.; Micheletti, G.; Porcu, M.; Mannelli, L.; Bassareo, P.; Suri, J.S.; Saba, L. Potential role of artificial intelligence in cardiac magnetic resonance imaging: Can it help clinicians in making a diagnosis? J. Thorac. Imaging 2021, 36, 142–148.
- Laine, A.; Sanches, J.M.; Suri, J.S. Ultrasound Imaging: Advances and Applications; Springer: Cham, Seitzerland, 2012.
- Radeva, P.; Suri, J.S. Vascular and Intravascular Imaging Trends, Analysis, and Challenges: Plaque Characterization; IOP Publishing: Bristol, UK, 2019; Volume 2.
- Sun, Z.; Xu, L. Coronary CT angiography in the quantitative assessment of coronary plaques. BioMed Res. Int. 2014, 2014, 346380.
- Cau, R.; Flanders, A.; Mannelli, L.; Politi, C.; Faa, G.; Suri, J.S.; Saba, L. Artificial intelligence in computed tomography plaque characterization: A review. Eur. J. Radiol. 2021, 140, 109767.
- Murgia, A.; Balestrieri, A.; Crivelli, P.; Suri, J.S.; Conti, M.; Cademartiri, F.; Saba, L. Cardiac computed tomography radiomics: An emerging tool for the non-invasive assessment of coronary atherosclerosis. Cardiovasc. Diagn. Ther. 2020, 10, 2005.
- Onnis, C.; Cadeddu Dessalvi, C.; Cademartiri, F.; Muscogiuri, G.; Angius, S.; Contini, F.; Suri, J.S.; Sironi, S.; Salgado, R.; Esposito, A. Quantitative and qualitative features of carotid and coronary atherosclerotic plaque among men and women. Front. Cardiovasc. Med. 2022, 9, 970438.
- Onnis, C.; Muscogiuri, G.; Bassareo, P.P.; Cau, R.; Mannelli, L.; Cadeddu, C.; Suri, J.S.; Cerrone, G.; Gerosa, C.; Sironi, S. Non-invasive coronary imaging in patients with COVID-19: A narrative review. Eur. J. Radiol. 2022, 149, 110188.
- Ghekiere, O.; Salgado, R.; Buls, N.; Leiner, T.; Mancini, I.; Vanhoenacker, P.; Dendale, P.; Nchimi, A. Image quality in coronary CT angiography: Challenges and technical solutions. Br. J. Radiol. 2017, 90, 20160567.
- Ozolanta, I.; Tetere, G.; Purinya, B.; Kasyanov, V. Changes in the mechanical properties, biochemical contents and wall structure of the human coronary arteries with age and sex. Med. Eng. Phys. 1998, 20, 523–533.
- Hayes, S.N.; Kim, E.S.; Saw, J.; Adlam, D.; Arslanian-Engoren, C.; Economy, K.E.; Ganesh, S.K.; Gulati, R.; Lindsay, M.E.; Mieres, J.H. Spontaneous coronary artery dissection: Current state of the science: A scientific statement from the American Heart Association. Circulation 2018, 137, e523–e557.
- Song, A.; Xu, L.; Wang, L.; Wang, B.; Yang, X.; Xu, B.; Yang, B.; Greenwald, S.E. Automatic Coronary Artery Segmentation of CCTA Images with an Efficient Feature-Fusion-and-Rectification 3D-UNet. IEEE J. Biomed. Health Inform. 2022, 26, 4044–4055.
- Schroeder, S.; Kopp, A.F.; Kuettner, A.; Burgstahler, C.; Herdeg, C.; Heuschmid, M.; Baumbach, A.; Claussen, C.D.; Karsch, K.R.; Seipel, L. Influence of heart rate on vessel visibility in noninvasive coronary angiography using new multislice computed tomography: Experience in 94 patients. Clin. Imaging 2002, 26, 106–111.
- Araki, T.; Ikeda, N.; Shukla, D.; Londhe, N.D.; Shrivastava, V.K.; Banchhor, S.K.; Saba, L.; Nicolaides, A.; Shafique, S.; Laird, J.R. A new method for IVUS-based coronary artery disease risk stratification: A link between coronary & carotid ultrasound plaque burdens. Comput. Methods Programs Biomed. 2016, 124, 161–179.
- Ono, M.; Kawashima, H.; Hara, H.; Gao, C.; Wang, R.; Kogame, N.; Takahashi, K.; Chichareon, P.; Modolo, R.; Tomaniak, M. Advances in IVUS/OCT and future clinical perspective of novel hybrid catheter system in coronary imaging. Front. Cardiovasc. Med. 2020, 7, 119.
- Faraji, M.; Cheng, I.; Naudin, I.; Basu, A. Segmentation of arterial walls in intravascular ultrasound cross-sectional images using extremal region selection. Ultrasonics 2018, 84, 356–365.
- Suri, J.S.; Paul, S.; Maindarkar, M.A.; Puvvula, A.; Saxena, S.; Saba, L.; Turk, M.; Laird, J.R.; Khanna, N.N.; Viskovic, K. Cardiovascular/stroke risk stratification in Parkinson’s disease patients using atherosclerosis pathway and artificial intelligence paradigm: A systematic review. Metabolites 2022, 12, 312.
- Jena, B.; Saxena, S.; Nayak, G.K.; Balestrieri, A.; Gupta, N.; Khanna, N.N.; Laird, J.R.; Kalra, M.K.; Fouda, M.M.; Saba, L.; et al. Brain Tumor Characterization Using Radiogenomics in Artificial Intelligence Framework. Cancers 2022, 14, 4052.
- Suri, J.S.; Maindarkar, M.A.; Paul, S.; Ahluwalia, P.; Bhagawati, M.; Saba, L.; Faa, G.; Saxena, S.; Singh, I.M.; Chadha, P.S. Deep Learning Paradigm for Cardiovascular Disease/Stroke Risk Stratification in Parkinson’s Disease Affected by COVID-19: A Narrative Review. Diagnostics 2022, 12, 1543.
- Suri, J.S.; Bhagawati, M.; Paul, S.; Protogerou, A.D.; Sfikakis, P.P.; Kitas, G.D.; Khanna, N.N.; Ruzsa, Z.; Sharma, A.M.; Saxena, S. A powerful paradigm for cardiovascular risk stratification using multiclass, multi-label, and ensemble-based machine learning paradigms: A narrative review. Diagnostics 2022, 12, 722.
- Das, S.; Nayak, G.K.; Saba, L.; Kalra, M.; Suri, J.S.; Saxena, S. An artificial intelligence framework and its bias for brain tumor segmentation: A narrative review. Comput. Biol. Med. 2022, 143, 105273.
- Suri, J.S.; Agarwal, S.; Jena, B.; Saxena, S.; El-Baz, A.; Agarwal, V.; Kalra, M.K.; Saba, L.; Viskovic, K.; Fatemi, M. Five strategies for bias estimation in artificial intelligence-based hybrid deep learning for acute respiratory distress syndrome COVID-19 lung infected patients using AP (AI) Bias 2.0: A systematic review. In IEEE Transactions on Instrumentation and Measurement; IEEE: Piscataway, NJ, USA, 2022.
- Suri, J.S.; Agarwal, S.; Gupta, S.K.; Puvvula, A.; Viskovic, K.; Suri, N.; Alizad, A.; El-Baz, A.; Saba, L.; Fatemi, M. Systematic review of artificial intelligence in acute respiratory distress syndrome for COVID-19 lung patients: A biomedical imaging perspective. IEEE J. Biomed. Health Inform. 2021, 25, 4128–4139.
- Ikeda, N.; Gupta, A.; Dey, N.; Bose, S.; Shafique, S.; Arak, T.; Godia, E.C.; Saba, L.; Laird, J.R.; Nicolaides, A. Improved correlation between carotid and coronary atherosclerosis SYNTAX score using automated ultrasound carotid bulb plaque IMT measurement. Ultrasound Med. Biol. 2015, 41, 1247–1262.
- Ikeda, N.; Araki, T.; Sugi, K.; Nakamura, M.; Deidda, M.; Molinari, F.; Meiburger, K.M.; Acharya, U.R.; Saba, L.; Bassareo, P.P. Ankle–brachial index and its link to automated carotid ultrasound measurement of intima–media thickness variability in 500 Japanese coronary artery disease patients. Curr. Atheroscler. Rep. 2014, 16, 1–8.
- Kumar, A.; Aelgani, V.; Vohra, R.; Gupta, S.K.; Bhagawati, M.; Paul, S.; Saba, L.; Suri, N.; Khanna, N.N.; Laird, J.R. Artificial intelligence bias in medical system designs: A systematic review. Multimedia Tools and Applications. 2023, 347, 1–53.
- Araki, T.; Banchhor, S.K.; Londhe, N.D.; Ikeda, N.; Radeva, P.; Shukla, D.; Saba, L.; Balestrieri, A.; Nicolaides, A.; Shafique, S. Reliable and accurate calcium volume measurement in coronary artery using intravascular ultrasound videos. J. Med. Syst. 2016, 40, 1–20.
- Suri, J.S. Computer vision, pattern recognition and image processing in left ventricle segmentation: The last 50 years. Pattern Anal. Appl. 2000, 3, 209–242.
- El-Baz, A.S.; Acharya, R.; Mirmehdi, M.; Suri, J.S. Multi Modality State-of-the-Art Medical Image Segmentation and Registration Methodologies; Springer Science & Business Media: Cham, Switzerland, 2011; Volume 1.
- El-Baz, A.; Jiang, X.; Suri, J.S. Biomedical Image Segmentation: Advances and Trends; CRC Press: Boca Raton, FL, USA, 2016; p. 1.
- El-Baz, A.; Suri, J.S. Level Set Method in Medical Imaging Segmentation; CRC Press: Boca Raton, FL, USA, 2019.
- Kumar, A.; Jain, R. Behavioral Prediction of Cancer Using Machine Learning; Chapman and Hall/CRC Press: Boca Raton, FL, USA, 2021; pp. 91–105.
- Bindu, C.H. An improved medical image segmentation algorithm using Otsu method. Int. J. Recent Trends Eng. 2009, 2, 88.
- Noor, N.M.; Than, J.C.; Rijal, O.M.; Kassim, R.M.; Yunus, A.; Zeki, A.A.; Anzidei, M.; Saba, L.; Suri, J.S. Automatic lung segmentation using control feedback system: Morphology and texture paradigm. J. Med. Syst. 2015, 39, 1–18.
- Suri, J.S. Leaking prevention in fast level sets using fuzzy models: An application in MR brain. In Proceedings of the 2000 IEEE EMBS International Conference on Information Technology Applications in Biomedicine. ITAB-ITIS 2000. Joint Meeting Third IEEE EMBS International Conference on Information Technol, Arlington, VA, USA, 9–10 November 2000; pp. 220–225.
- Suri, J.S.; Liu, K. Level set regularizers for shape recovery in medical images. In Proceedings of the 14th IEEE Symposium on Computer-Based Medical Systems. CBMS 2001, Bethesda, MD, USA, 26–27 July 2001; pp. 369–374.
- Jain, P.K.; Sharma, N.; Saba, L.; Paraskevas, K.I.; Kalra, M.K.; Johri, A.; Laird, J.R.; Nicolaides, A.N.; Suri, J.S. Unseen artificial intelligence—Deep learning paradigm for segmentation of low atherosclerotic plaque in carotid ultrasound: A multicenter cardiovascular study. Diagnostics 2021, 11, 2257.
- Paul, S.; Maindarkar, M.; Saxena, S.; Saba, L.; Turk, M.; Kalra, M.; Krishnan, P.R.; Suri, J.S. Bias investigation in artificial intelligence systems for early detection of Parkinson’s disease: A narrative review. Diagnostics 2022, 12, 166.
- Suri, J.S.; Agarwal, S.; Chabert, G.L.; Carriero, A.; Paschè, A.; Danna, P.S.; Saba, L.; Mehmedović, A.; Faa, G.; Singh, I.M. COVLIAS 1.0 Lesion vs. MedSeg: An Artificial Intelligence Framework for Automated Lesion Segmentation in COVID-19 Lung Computed Tomography Scans. Diagnostics 2022, 12, 1283.
- Suri, J.S.; Agarwal, S.; Chabert, G.L.; Carriero, A.; Paschè, A.; Danna, P.S.; Saba, L.; Mehmedović, A.; Faa, G.; Singh, I.M. COVLIAS 2.0-cXAI: Cloud-based explainable deep learning system for COVID-19 lesion localization in computed tomography scans. Diagnostics 2022, 12, 1482.
- Sharma, N.; Saba, L.; Khanna, N.N.; Kalra, M.K.; Fouda, M.M.; Suri, J.S. Segmentation-Based Classification Deep Learning Model Embedded with Explainable AI for COVID-19 Detection in Chest X-ray Scans. Diagnostics 2022, 12, 2132.
- El-Baz, A.S.; Suri, J.S. State of the Art in Neural Networks and Their Applications; Volume 1: Imaging and Signal Analysis; Academic Press: Cambridge, MA, USA, 2021.
- Kumar, K.; Saeed, U.; Rai, A.; Islam, N.; Shaikh, G.M.; Qayoom, A. Idc breast cancer detection using deep learning schemes. Adv. Data Sci. Adapt. Anal. 2020, 12, 2041002.
- Saba, L.; Biswas, M.; Kuppili, V.; Godia, E.C.; Suri, H.S.; Edla, D.R.; Omerzu, T.; Laird, J.R.; Khanna, N.N.; Mavrogeni, S. The present and future of deep learning in radiology. Eur. J. Radiol. 2019, 114, 14–24.
- Dubey, A.K.; Chabert, G.L.; Carriero, A.; Pasche, A.; Danna, P.S.; Agarwal, S.; Mohanty, L.; Nillmani; Sharma, N.; Yadav, S. Ensemble Deep Learning Derived from Transfer Learning for Classification of COVID-19 Patients on Hybrid Deep-Learning-Based Lung Segmentation: A Data Augmentation and Balancing Framework. Diagnostics 2023, 13, 1954.
- He, X.; Guo, B.J.; Lei, Y.; Wang, T.; Fu, Y.; Curran, W.J.; Zhang, L.J.; Liu, T.; Yang, X. Automatic segmentation and quantification of epicardial adipose tissue from coronary computed tomography angiography. Phys. Med. Biol. 2020, 65, 095012.
- Jin, X.; Li, Y.; Yan, F.; Liu, Y.; Zhang, X.; Li, T.; Yang, L.; Chen, H. Automatic coronary plaque detection, classification, and stenosis grading using deep learning and radiomics on computed tomography angiography images: A multi-center multi-vendor study. Eur. Radiol. 2022, 32, 5276–5286.
- Ibtehaz, N.; Rahman, M.S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87.
- Balakrishna, C.; Dadashzadeh, S.; Soltaninejad, S. Automatic detection of lumen and media in the IVUS images using U-Net with VGG16 Encoder. arXiv 2018, arXiv:1806.07554.
- Kim, S.; Jang, Y.; Jeon, B.; Hong, Y.; Shim, H.; Chang, H. Fully automatic segmentation of coronary arteries based on deep neural network in intravascular ultrasound images. In Intravascular Imaging and Computer Assisted Stenting and Large-Scale Annotation of Biomedical Data and Expert Label Synthesis; 7th Joint International Workshop, CVII-STENT 2018 and Third International Workshop, LABELS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 16, 2018, Proceedings 3; Springer International Publishing: Cham, Switzerland, 2018; pp. 161–168.
- Li, Y.-C.; Shen, T.-Y.; Chen, C.-C.; Chang, W.-T.; Lee, P.-Y.; Huang, C.-C.J. Automatic detection of atherosclerotic plaque and calcification from intravascular ultrasound images by using deep convolutional neural networks. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 2021, 68, 1762–1772.
- Chen, Y.-C.; Lin, Y.-C.; Wang, C.-P.; Lee, C.-Y.; Lee, W.-J.; Wang, T.-D.; Chen, C.-M. Coronary artery segmentation in cardiac CT angiography using 3D multi-channel U-net. arXiv 2019, arXiv:1907.12246.
- Tong, Q.; Ning, M.; Si, W.; Liao, X.; Qin, J. 3D deeply-supervised U-net based whole heart segmentation. In Statistical Atlases and Computational Models of the Heart. ACDC and MMWHS Challenges: 8th International Workshop, STACOM 2017, Held in Conjunction with MICCAI 2017, Quebec City, Canada, September 10–14, 2017, Revised Selected Papers 8; Springer International Publishing: Cham, Switzerland, 2018; pp. 224–232.
- Morris, E.D.; Ghanem, A.I.; Dong, M.; Pantelic, M.V.; Walker, E.M.; Glide-Hurst, C.K. Cardiac substructure segmentation with deep learning for improved cardiac sparing. Med. Phys. 2020, 47, 576–586.
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imaging 2019, 39, 1856–1867.
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571.
- Szarski, M.; Chauhan, S. Improved real-time segmentation of Intravascular Ultrasound images using coordinate-aware fully convolutional networks. Comput. Med. Imaging Graph. 2021, 91, 101955.
- Vercio, L.L.; Del Fresno, M.; Larrabide, I. Lumen-intima and media-adventitia segmentation in IVUS images using supervised classifications of arterial layers and morphological structures. Comput. Methods Programs Biomed. 2019, 177, 113–121.
- Yang, J.; Faraji, M.; Basu, A. Robust segmentation of arterial walls in intravascular ultrasound images using Dual Path U-Net. Ultrasonics 2019, 96, 24–33.
- Shen, Y.; Fang, Z.; Gao, Y.; Xiong, N.; Zhong, C.; Tang, X. Coronary arteries segmentation based on 3D FCN with attention gate and level set function. IEEE Access 2019, 7, 42826–42835.
- Jávorszky, N.; Homonnay, B.; Gerstenblith, G.; Bluemke, D.; Kiss, P.; Török, M.; Celentano, D.; Lai, H.; Lai, S.; Kolossváry, M. Deep learning–based atherosclerotic coronary plaque segmentation on coronary CT angiography. Eur. Radiol. 2022, 32, 7217–7226.
- Momin, S.; Lei, Y.; McCall, N.S.; Zhang, J.; Roper, J.; Harms, J.; Tian, S.; Lloyd, M.S.; Liu, T.; Bradley, J.D. Mutual enhancing learning-based automatic segmentation of CT cardiac substructure. Phys. Med. Biol. 2022, 67, 105008.
- Guo, C.; Li, P. Hybrid Pruning Method Based on Convolutional Neural Network Sensitivity and Statistical Threshold. In Journal of Physics: Conference Series; IOP Publishing: Bristol, UK, 2022; Volume 2171, p. 012055.
- Huang, C.; Lan, Y.; Xu, G.; Zhai, X.; Wu, J.; Lin, F.; Zeng, N.; Hong, Q.; Ng, E.; Peng, Y. A deep segmentation network of multi-scale feature fusion based on attention mechanism for IVOCT lumen contour. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 18, 62–69.
- Jun, T.J.; Kweon, J.; Kim, Y.-H.; Kim, D. T-net: Nested encoder–decoder architecture for the main vessel segmentation in coronary angiography. Neural Netw. 2020, 128, 216–233.
- Shi, X.; Du, T.; Chen, S.; Zhang, H.; Guan, C.; Xu, B. UENet: A novel generative adversarial network for angiography image segmentation. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1612–1615.
- Thuy, L.N.L.; Trinh, T.D.; Anh, L.H.; Kim, J.Y.; Hieu, H.T. Coronary vessel segmentation by coarse-to-fine strategy using u-nets. BioMed Res. Int. 2021, 2021, 5548517.
- Hwang, M.; Hwang, S.-B.; Yu, H.; Kim, J.; Kim, D.; Hong, W.; Ryu, A.-J.; Cho, H.Y.; Zhang, J.; Koo, B.K. A Simple Method for Automatic 3D Reconstruction of Coronary Arteries from X-ray Angiography. Front. Physiol. 2021, 12, 724216.
- Cheung, W.K.; Bell, R.; Nair, A.; Menezes, L.J.; Patel, R.; Wan, S.; Chou, K.; Chen, J.; Torii, R.; Davies, R.H. A computationally efficient approach to segmentation of the aorta and coronary arteries using deep learning. IEEE Access 2021, 9, 108873–108888.
- Dong, L.; Jiang, W.; Lu, W.; Jiang, J.; Zhao, Y.; Song, X.; Leng, X.; Zhao, H.; Wang, J.A.; Li, C. Automatic segmentation of coronary lumen and external elastic membrane in intravascular ultrasound images using 8-layer U-Net. BioMedical Eng. OnLine 2021, 20, 1–9.
- Pan, L.-S.; Li, C.-W.; Su, S.-F.; Tay, S.-Y.; Tran, Q.-V.; Chan, W.P. Coronary artery segmentation under class imbalance using a U-Net based architecture on computed tomography angiography images. Sci. Rep. 2021, 11, 14493.
- Shinohara, H.; Kodera, S.; Ninomiya, K.; Nakamoto, M.; Katsushika, S.; Saito, A.; Minatsuki, S.; Kikuchi, H.; Kiyosue, A.; Higashikuni, Y. Automatic detection of vessel structure by deep learning using intravascular ultrasound images of the coronary arteries. PLoS ONE 2021, 16, e0255577.
- Xia, M.; Yan, W.; Huang, Y.; Guo, Y.; Zhou, G.; Wang, Y. Extracting membrane borders in ivus images using a multi-scale feature aggregated u-net. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1650–1653.
- Azad, R.; Asadi-Aghbolaghi, M.; Fathy, M.; Escalera, S. Bi-directional ConvLSTM U-Net with densley connected convolutions. In Proceedings of the IEEE/CVF international conference on computer vision workshops, Montreal, BC, Canada, 11–17 October 2021.
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5–9, 2015, Proceedings, Part III 18 (pp. 234–241); Springer International Publishing: Cham, Switzerland, 2015.
- Bajaj, R.; Huang, X.; Kilic, Y.; Ramasamy, A.; Jain, A.; Ozkor, M.; Tufaro, V.; Safi, H.; Erdogan, E.; Serruys, P.W. Advanced deep learning methodology for accurate, real-time segmentation of high-resolution intravascular ultrasound images. Int. J. Cardiol. 2021, 339, 185–191.
- Cho, H.; Kang, S.-J.; Min, H.-S.; Lee, J.-G.; Kim, W.-J.; Kang, S.H.; Kang, D.-Y.; Lee, P.H.; Ahn, J.-M.; Park, D.-W. Intravascular ultrasound-based deep learning for plaque characterization in coronary artery disease. Atherosclerosis 2021, 324, 69–75.
- Bajaj, R.; Huang, X.; Kilic, Y.; Jain, A.; Ramasamy, A.; Torii, R.; Moon, J.; Koh, T.; Crake, T.; Parker, M.K. A deep learning methodology for the automated detection of end-diastolic frames in intravascular ultrasound images. Int. J. Cardiovasc. Imaging 2021, 37, 1825–1837.
- Fedewa, R.; Puri, R.; Fleischman, E.; Lee, J.; Prabhu, D.; Wilson, D.L.; Vince, D.G.; Fleischman, A. Artificial intelligence in intracoronary imaging. Curr. Cardiol. Rep. 2020, 22, 1–15.
- Masuda, T.; Nakaura, T.; Funama, Y.; Oda, S.; Okimoto, T.; Sato, T.; Noda, N.; Yoshiura, T.; Baba, Y.; Arao, S. Deep learning with convolutional neural network for estimation of the characterisation of coronary plaques: Validation using IB-IVUS. Radiography 2022, 28, 61–67.
- Min, H.-S.; Ryu, D.; Kang, S.-J.; Lee, J.-G.; Yoo, J.H.; Cho, H.; Kang, D.-Y.; Lee, P.H.; Ahn, J.-M.; Park, D.-W. Prediction of coronary stent underexpansion by pre-procedural intravascular ultrasound–based deep learning. Cardiovasc. Interv. 2021, 14, 1021–1029.
- Nishi, T.; Yamashita, R.; Imura, S.; Tateishi, K.; Kitahara, H.; Kobayashi, Y.; Yock, P.G.; Fitzgerald, P.J.; Honda, Y. Deep learning-based intravascular ultrasound segmentation for the assessment of coronary artery disease. Int. J. Cardiol. 2021, 333, 55–59.
- Shin, C.-I.; Park, S.J.; Kim, J.-H.; Yoon, Y.E.; Park, E.-A.; Koo, B.-K.; Lee, W. Coronary Artery Lumen Segmentation Using Location–Adaptive Threshold in Coronary Computed Tomographic Angiography: A Proof-of-Concept. Korean J. Radiol. 2021, 22, 688.
- Olender, M.L.; Athanasiou, L.S.; Michalis, L.K.; Fotiadis, D.I.; Edelman, E.R. A domain enriched deep learning approach to classify atherosclerosis using intravascular ultrasound imaging. IEEE J. Sel. Top. Signal Process. 2020, 14, 1210–1220.
- Zhao, F.; Wu, B.; Chen, F.; Cao, X.; Yi, H.; Hou, Y.; He, X.; Liang, J. An automatic multi-class coronary atherosclerosis plaque detection and classification framework. Med. Biol. Eng. Comput. 2019, 57, 245–257.
- Bargsten, L.; Raschka, S.; Schlaefer, A. Capsule networks for segmentation of small intravascular ultrasound image datasets. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 1243–1254.
- Samuel, P.M.; Veeramalai, T. VSSC Net: Vessel specific skip chain convolutional network for blood vessel segmentation. Comput. Methods Programs Biomed. 2021, 198, 105769.
- Tayel, M.B.; Massoud, M.; Farouk, Y. A modified segmentation method for determination of IV vessel boundaries. Alex. Eng. J. 2017, 56, 449–457.
- Cui, H.; Xia, Y.; Zhang, Y. Supervised machine learning for coronary artery lumen segmentation in intravascular ultrasound images. Int. J. Numer. Methods Biomed. Eng. 2020, 36, e3348.
- Harms, J.; Lei, Y.; Tian, S.; McCall, N.S.; Higgins, K.A.; Bradley, J.D.; Curran, W.J.; Liu, T.; Yang, X. Automatic delineation of cardiac substructures using a region-based fully convolutional network. Med. Phys. 2021, 48, 2867–2876.
- Mishra, D.; Chaudhury, S.; Sarkar, M.; Soin, A.S. Ultrasound image segmentation: A deeply supervised network with attention to boundaries. IEEE Trans. Biomed. Eng. 2018, 66, 1637–1648.
- Lin, A.; Manral, N.; McElhinney, P.; Killekar, A.; Matsumoto, H.; Kwiecinski, J.; Pieszko, K.; Razipour, A.; Grodecki, K.; Park, C. Deep learning-enabled coronary CT angiography for plaque and stenosis quantification and cardiac risk prediction: An international multicentre study. Lancet Digit. Health 2022, 4, e256–e265.
- Du, H.; Ling, L.; Yu, W.; Wu, P.; Yang, Y.; Chu, M.; Yang, J.; Yang, W.; Tu, S. Convolutional networks for the segmentation of intravascular ultrasound images: Evaluation on a multicenter dataset. Comput. Methods Programs Biomed. 2022, 215, 106599.
- Jodas, D.S.; Pereira, A.S.; Tavares, J.M.R. Automatic segmentation of the lumen region in intravascular images of the coronary artery. Med. Image Anal. 2017, 40, 60–79.
- Eslamizadeh, M.; Attarodi, G.; Dabanloo, N.J.; Sedehi, J.F.; Setaredan, S.K. The segmentation of lumen boundaries at intravascular ultrasound images using fuzzy approach. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017; pp. 1–4.
- Sofian, H.; Than, J.C.; Noor, N.M.; Dao, H. Segmentation and detection of media adventitia coronary artery boundary in medical imaging intravascular ultrasound using otsu thresholding. In Proceedings of the 2015 International Conference on BioSignal Analysis, Processing and Systems (ICBAPS), Kuala Lumpur, Malaysia, 26–28 May 2015; pp. 72–76.
- Cao, Y.; Wang, Z.; Liu, Z.; Li, Y.; Xiao, X.; Sun, L.; Zhang, Y.; Hou, H.; Zhang, P.; Yang, G. Multiparameter synchronous measurement with IVUS images for intelligently diagnosing coronary cardiac disease. IEEE Trans. Instrum. Meas. 2020, 70, 1–10.
- Taki, A.; Najafi, Z.; Roodaki, A.; Setarehdan, S.K.; Zoroofi, R.A.; Konig, A.; Navab, N. Automatic segmentation of calcified plaques and vessel borders in IVUS images. Int. J. Comput. Assist. Radiol. Surg. 2008, 3, 347–354.
- Unal, G.; Bucher, S.; Carlier, S.; Slabaugh, G.; Fang, T.; Tanaka, K. Shape-driven segmentation of the arterial wall in intravascular ultrasound images. IEEE Trans. Inf. Technol. Biomed. 2008, 12, 335–347.
- Zhu, X.; Zhang, P.; Shao, J.; Cheng, Y.; Zhang, Y.; Bai, J. A snake-based method for segmentation of intravascular ultrasound images and its in vivo validation. Ultrasonics 2011, 51, 181–189.
- Sanagala, S.S.; Nicolaides, A.; Gupta, S.K.; Koppula, V.K.; Saba, L.; Agarwal, S.; Johri, A.M.; Kalra, M.S.; Suri, J.S. Ten fast transfer learning models for carotid ultrasound plaque tissue characterization in augmentation framework embedded with heatmaps for stroke risk stratification. Diagnostics 2021, 11, 2109.
- Jain, P.K.; Sharma, N.; Giannopoulos, A.A.; Saba, L.; Nicolaides, A.; Suri, J.S. Hybrid deep learning segmentation models for atherosclerotic plaque in internal carotid artery B-mode ultrasound. Comput. Biol. Med. 2021, 136, 104721.
- Skandha, S.S.; Gupta, S.K.; Saba, L.; Koppula, V.K.; Johri, A.M.; Khanna, N.N.; Mavrogeni, S.; Laird, J.R.; Pareek, G.; Miner, M. 3-D optimized classification and characterization artificial intelligence paradigm for cardiovascular/stroke risk stratification using carotid ultrasound-based delineated plaque: Atheromatic™ 2.0. Comput. Biol. Med. 2020, 125, 103958.
- Agarwal, M.; Saba, L.; Gupta, S.K.; Johri, A.M.; Khanna, N.N.; Mavrogeni, S.; Laird, J.R.; Pareek, G.; Miner, M.; Sfikakis, P.P. Wilson disease tissue classification and characterization using seven artificial intelligence models embedded with 3D optimization paradigm on a weak training brain magnetic resonance imaging datasets: A supercomputer application. Med. Biol. Eng. Comput. 2021, 59, 511–533.
More
