Candidiasis is a highly pervasive infection posing major health risks, especially for immunocompromised populations. Pathogenic Candida species have evolved intrinsic and acquired resistance to a variety of antifungal medications.
- antifungal
- resistance
- Candida
1. Introduction
1.1. Candidiasis
1.1.1. Candida Species of Interest
Phylogenetic categorization of Candida yeast suggests polyphyly and pathogenic diversity among members [18][13]. Three of these species (C. albicans, C. parapsilosis and C. tropicalis) belong to the CTG clade, which contains most pathogenic Candida species, while P. kudriavzevii is more closely related to a wine-making yeast (Brettanomyces bruxellensis) [18][13]. Members of the CTG clade have a divergence in their genetic code compared to other Saccharomycotina subphylum yeast [19,20][14][15]. These species are categorized based on the CTG codon being transcribed and translated into serine instead of a typical leucine [19,20][14][15]. C. glabrata is part of the Nakaseomyces clade and was recently renamed Nakaseomyces glabrata for improved classification [21][16]. Despite this species being one of the few pathogenic members of its clade, it is the second most common cause of candidiasis globally [18][13]. The consequences of genetic alterations in N. glabrata may diverge from typical Candida species because it is a haploid organism [22][17]. Resistance could arise at a higher rate because a single recessive point mutation can present phenotypically due to a haploid genome. This contrasts with other Candida species that are diploid and therefore may require two copies of the mutated gene to present with resistance [23][18].1.1.2. Candida auris
Candida auris of the CTG clade is listed under critical priority in the FPPL due to its high infectiousness, global spread and high fatality risk [17,18,24,25][13][19][20][21]. Numerous species isolates have been identified as displaying resistance to several antifungals [26,27,28][22][23][24]. An update on C. auris released by Public Health Ontario (2023) indicated high rates of resistance to azole drug fluconazole (87–100%), while polyene amphotericin B and echinocandin resistances are cited less frequently, with ranges of 8–35% and 0–8%, respectively [29][25]. The CDC reported a similar rate of approximately 30% for polyene-resistant strains [30][26]. At least 4% of global cases of C. auris infections display multidrug resistance to all three antifungal types, which can make adequate clinical treatment especially difficult [29][25].1.2. Primary/Intrinsic Resistance vs. Secondary/Acquired Resistance
Fungal resistance can be divided into primary/intrinsic and secondary/acquired resistance. Some fungal species are intrinsically resistant to a specific antifungal drug because of innate functional or structural attributes. This stable feature is seen in all strains from the same species and has not evolved due to previous antifungal exposure [1]. One example is the intrinsically fluconazole-resistant P. kudriavzevii [32,33][27][28]. Alternatively, acquired resistance can evolve in strains of a Candida species that are typically susceptible to an antifungals. This secondary form of resistance usually develops after prolonged treatment in a clinical or in vitro setting [34][29]. Mutations or chromosomal rearrangements can cause an overexpression of genes that override the effects of antifungal activity or the fungal stress response [34][29]. This change can revert to the original state once the pressure of the drug treatment is reduced or removed. Some mutants may retain the resistant phenotype regardless of future drug pressure [34][29]. Antifungal action can be evaded by pathogenic yeast by two main methods: (1) An alteration of the interaction between drug and target. This can result from either a change in the target protein amino acid (aa) sequence and, consequently, its structure or target protein overexpression. Alternatively, (2) the cytoplasmic drug concentration can be reduced via cell wall modifications that decrease drug absorption into the cell or the overexpression of efflux pumps that promote the export of drug molecules out of the cell [34][29]. Multidrug resistance (MDR) occurs when these mutations accumulate in the yeast genome in target pathways, which limits the amount of treatment options available [23,35][18][30]. An example of MDR was observed in C. albicans isolates with resistance to both fluconazole and clotrimazole [36][31]. Detecting mutant genotypes with acquired resistance in a timely manner could be an independent and useful predictive risk factor for treatment failure [34,37,38][29][32][33].1.3. Standardized Measures of Susceptibility Testing
Minimum inhibitory concentrations (MICs) calculated with in vitro broth microdilution susceptibility testing are used to categorize dose-dependent resistance in Candida species [39,40,41][34][35][36]. There are two main standardization methods that outline the established breakpoint concentrations for ranges of resistance: those of the Clinical Standards Laboratory Institute (CLSI—North America) and the European Committee on Antimicrobial Testing (EUCAST) [40,41,42][35][36][37]. Considering that C. parapsilosis cryptic species C. orthopsilosis and C. metapsilosis are of low prevalence, the MIC breakpoint data for C. parapsilosis can be applied in cases when further species characterization has not been completed [43,44,45,46,47][38][39][40][41][42]. One trend identified in the data is the tendency of high-resistance concentrations in N. glabrata for fluconazole, whereas lower MICs are implicated with the use of echinocandins for this species.2. Antifungal Classes and Frequency of Resistance
A range of antifungal drug classes is available to target various molecules and pathways associated with pathogenic Candida infections. The reviews by Bhattacharya et al. (2020) and Tilley and Tharmalingam (2022) provide an excellent summary of the four primary antifungal drug classes: azoles, polyenes, echinocandins and nucleoside analogs [1,55][1][43]. The section below summarizes the mechanisms of action for each antifungal drug class (Figure 1), as well as relevant resistance profiles. The molecular structures of drugs from each antifungal class and key points for each type are presented in Figure 2.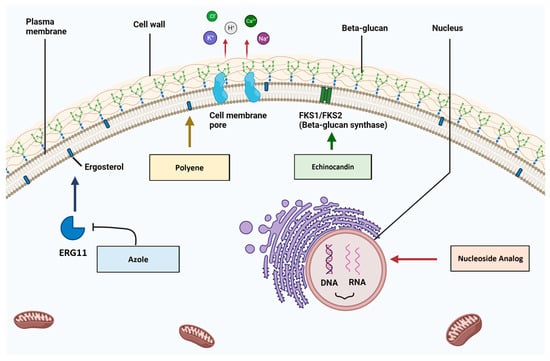
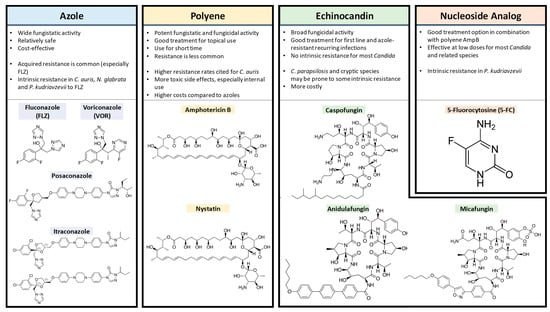
2.1. Azoles
2.2. Polyenes
Polyene antifungals like amphotericin B and nystatin target the fungal plasma membrane by binding ergosterol molecules and forming pores that leak cell contents (monovalent ions K+, Na+, H+ and Cl−) [67][49]. These are potent agents with fungicidal and fungistatic activity that are used in clinical practice for their effectiveness despite relatively higher rates of toxic side effects like kidney/liver issues or anaphylaxis [50,68,69][50][51][52]. Fungicidal agents can kill infectious yeast cells directly [1]. To limit the possibility of treatment toxicity, this class is best used for topical infections such as in the oral cavity and for a limited time course [69][52]. Reports of isolated fungal strains with acquired polyene resistance are relatively rare, despite decades of use in the clinical setting [70,71][53][54]. This may be attributed to the effectiveness of their fungicidal activity in eliminating infections and thus preventing the evolution of stable resistant mutants.2.3. Echinocandins
Echinocandins including caspofungin, micafungin and anidulafungin target the fungal cell wall, a feature not found in mammalian cells [73][55]. Echinocandins are composed of cyclic hexapeptides with lipid side-chain modifications that enable antifungal action [74][56]. They inhibit the synthesis of a major cell wall component via non-competitive binding to the Fks1 subunit of the β1–3 glucan synthase enzyme [75][57]. This action promotes a fungicidal effect, as the cell wall integrity is compromised, with increased permeability and subsequent amino acid leakage [74][56]. C. albicans tends to be the most susceptible to caspofungin, followed by N. glabrata, C. tropicalis, P. kudriavzevii, C. parapsilosis and M. guilliermondii [74][56]. The last two species listed seem to have more naturally arising FKS1 point mutations; thus, C. parapsilosis and M. guilliermondii appear to be more intrinsically echinocandin-resistant [77,78][58][59]. Secondary resistance to echinocandins has been observed and linked to point mutations in the FKS1 gene that alter antifungal binding capacity [79][60]. Strain viability may be compromised as indicated by the reduced virulence seen in multiple echinocandin-resistant Candida species [66][61].2.4. 5FC
5-Fluorocytosine (5FC) can be used to target and disrupt nucleic acid biosynthesis within the cell [80][62]. This nucleoside analog used in conjunction with polyene amphotericin B is a reliable option for difficult-to-treat Candida infections and cryptococcal meningitis [81,82,83][63][64][65]. The minimum inhibitory concentration for 90% of fungal growth (MIC90) (National Committee for Clinical Laboratory Standards (NCCLS)) determined using the antifungal susceptibility testing method has been cited from 0.12 to 1 ug/mL depending on the species and sample [83][65].3. The Ergosterol Biosynthesis Pathway and Antifungal Resistance
Azoles and polyenes target the ergosterol biosynthesis pathway, specifically the 14α–demethylase enzyme (Erg11) and ergosterol molecules respectively (Figure 3) [1]. Sterols, along with sphingolipids, can form lipid rafts within the fungal cell membrane that contain proteins crucial for yeast survival, like stress response, signaling and nutrient transport proteins [1]. There are 25 different pathway enzymes involved in the formation of ergosterol [85][66]. Azoles primarily exhibit fungistatic action via non-competitive binding to the Erg11 enzymatic active site, which inhibits its activity and results in an overall decrease in cellular levels of ergosterol [86][67].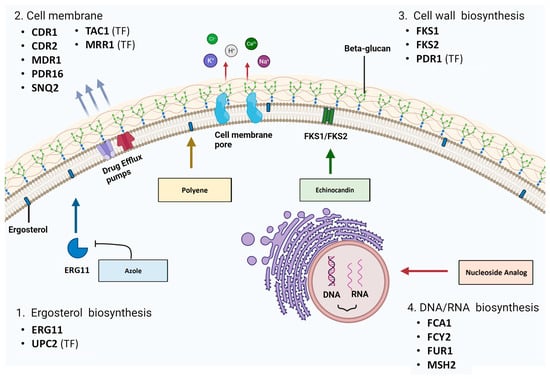
3.1. ERG11
By 2010, over 160 amino acid substitutions had been identified in the ERG11 gene, each with varying genetic consequences [87,88,89][68][69][70]. Many single substitutions are synonymous and have no impact on gene function. In addition, non-synonymous single-nucleotide changes occurring in Candida strains do not inherently contribute to antifungal resistance. For example, White et al. (2002) identified D116E and E266D, the two most frequent ERG11 substitutions in one sample set, using restriction fragment length polymorphism (RFLP) analysis, with no consistent correlation [36][31]. These instances indicate that there is natural genetic variation within each fungal species and that allelic polymorphisms are relatively common. Mutations present in both resistant and susceptible samples are an indicator that the alteration is not directly implicated in conferring antifungal resistance [86][67]. Candida spp. have acquired azole resistance via ERG11 point mutations that typically lower azole binding affinity to the Erg11 active site. Additionally, gain-of-function mutations in upstream transcriptional regulators can increase ERG11 expression and confer resistance. [34,86][29][67]. Point mutations resulting in a defective Erg11 enzyme unable to bind to azoles have been clustered in three hotspot regions: 105–165, 266–287 and 405–488 [94][71]. Polymorphisms can have different biological impacts, depending on whether they are singly present or in combination with other relevant SNPs. Resistance levels can be enhanced when some mutations with moderately low impact alone are present simultaneously with other resistance mutations [86,103,104][67][72][73]. The variety of currently available azole medications have different structural features (e.g., short and long chains); therefore, mutations affecting their efficacy may differ. For example, ERG11 point mutations K128T and Y132H may affect the ability of fluconazole or voriconazole molecules to enter or bind the target active site. Mutations in other gene sequences can also confer resistance, such as the G464S mutation, which affects haem coordination due to its location near a key cysteine residue [93][74]. These mutations do not have the same binding inefficiency for posaconazole and itraconazole treatments, suggesting that these two antifungals have other key interaction sites within the Erg11 protein [93][74].3.2. Mutations in Transcriptional Regulators
Zn2-Cys6 transcription factor uptake control 2 (Upc2) regulates most of the genes in the ergosterol biosynthesis pathway on some level. Gene overexpression of UPC2 can be induced upon azole exposure and can sufficiently compensate for the inhibition of target enzymes. Gain-of-function (GOF) mutations in UPC2 can drive this gene overexpression and fluconazole resistance [101][75]. For example, a series of studies of azole-resistant clinical isolates identified the A643V substitution in the UPC2 gene, which was validated in vitro to confer resistance [101,107][75][76]. This mutation and possibly others within this gene sequence region (G648D) may cause Upc2 to be released from a repressor, inducing hyperactivity [107][76]. However, alternate models exist that involve the sterol regulator, SREBP [107][76]. A genome-wide ChIP (chromatin immunoprecipitation) study used to identify Upc2-bound gene promoters identified up to 202 genes, including UPC2 itself [111][77]. Other upregulated genes were found to be involved in ergosterol biosynthesis; oxidoreductase activity; and numerous drug efflux pumps, including MDR1 (MFS-transporter) and CDR1 (ABC-transporter) [101][75].3.3. Other ERG Genes and Toxic Diol Formation
The inhibition of Erg11 alters pathway products and induces the synthesis of a fungistatic toxic diol (14α-methylergosta 8–24 (28) dienol) by downstream enzymes (Erg3, Erg6, Erg25, Erg26 and Erg27) [1]. Erg3 is a C5 sterol desaturase enzyme needed for the conversion of episterol to ergostatrienol [112][78]. When its expression is inhibited by mutation or deletion, the reduction in toxicity due to the inhibition of toxic diol formation is sufficient to confer resistance in some Candida spp. [113,114,115][79][80][81]. However, ERG3 inactivation appeared to minimally contribute to azole resistance in a wide range of studied clinical C. albicans [116][82]. Q139A substitution in ERG3 has been identified from N. glabrata clinical isolates with azole resistance [99][83]. Deletion of ERG pathway genes such as the ERG6 gene can impact resistance to other antifungals. This ∆24 sterol C-methyl transferase is non-essential for ergosterol biosynthesis but it is needed for toxic diol formation. Its disruption contributes to azole resistance in C. albicans [117,118][84][85]. One differential gene expression analysis of a lab-generated C. albicans strain with resistance to fluconazole and amphotericin B identified numerous upregulated ergosterol pathway genes, including ERG5, ERG6 and ERG25 [112][78]. Additionally, genes involved in cell stress responses were found to be upregulated, including DDR48 and RTA2 [112][78]. In C. albicans, RTA2 is a key gene in calcium signaling pathways, and it has been shown to modulate azole resistance, including biofilms [120][86].4. Cell Membrane Proteins and Antifungal Resistance
Two types of membrane transporters have been implicated in azole resistance: ABC-Ts (ATP-binding cassette transporters) and MFS-Transporters (major facilitator superfamily transporters) [36,110,121,122][31][87][88][89]. ABC-Ts facilitate the movement of molecules across membranes using energy derived from ATP hydrolysis, while MFS-Ts require a proton gradient across the plasma membrane to transport foreign molecules out of the cell [1]. Both types of transporters can bind azoles as a substrate, and the drug can be exported out of the cell. This decreases the intracellular drug concentration and allows cells to circumvent the antifungal effects [1]. An additional MLT1 (ABC-T) transporter has been implicated in C. albicans resistance. This multidrug resistance protein (MRP) is localized to the vacuolar membrane and can import azole molecules into the vacuole for sequestration. Mutations in the MLT1 sequence can cause incorrect localization or the inability to bind and transport azoles [123][90].4.1. Drug Efflux Pump/Transporter Genes and Resistant Mutations
Despite the large number of transporters found in the C. albicans genome, evidence of transporter overexpression in resistant clinical isolates is currently limited to ABC-Ts CDR1 and CDR2 and MFS-Ts MDR1 and PDR16 [1,111,131,133][1][77][91][92]. CDR1 and CDR2 overexpression is frequently observed in clinical isolates, and coregulation of these two pumps is evident [36][31]. In fact, prolonged exposure to azoles can result in trisomy development of chromosome 3, which encodes CDR1 and CDR2, as well as subsequent overexpression of these genes [124][93]. For azole-resistant N. glabrata clinical isolates, overexpression of ABC-T genes CDR1, PDH1 (CDR2 in C. albicans) and SNQ2 and MFS-T genes QDR2, FLR1 and PDR16 has been cited [62,104,134,135,136,137,138,139][73][94][95][96][97][98][99][100].4.2. Transcriptional Regulators of Transporter Genes
Efflux pump overexpression can be further induced by upstream GOF mutations, regulating transcription factor genes [59,125,126,127,128,129,130,144,145][47][101][102][103][104][105][106][107][108]. In C. albicans, the CDR1, CDR2 and PDR16 transporters, as well as MDR1, are regulated by zinc-cluster (Zn2-Cys6) transcription factors Tac1 and Mrr1, respectively [111,121,131,133,144,146][77][88][91][92][107][109]. Other potential transcriptional regulators of CDR1 expression include Tup1 (thymidine uptake 1) and Ncb2 (β subunit of the NC2 complex) [147][110]. For MDR1, additional transcriptional regulators are Cap1 (bZIP transcription factor) and Mcm1, but no antifungal-resistant mutations have been identified to date [148,149][111][112].4.3. Post-Translational Regulation of Transporter Genes
In addition to transcriptional and translational control of efflux pump genes, there is evidence of post-translational regulation. For instance, mitochondrial biogenesis gene FZO1 is important for directing Cdr1 to the correct membrane [153][113]. In FZO1 deletion mutants, Cdr1 was found to be mis-sorted to the vacuole, which was correlated with increased azole susceptibility [153][113].5. The Cell Wall Biosynthesis Pathway and Antifungal Resistance
5.1. FKS1 and FKS2 Sequence Mutations
Echinocandin resistance is associated with modifications to the FKS1 or FKS2 gene sequences, which code for the β1–3 glucan synthase enzyme [156][114]. FKS1 mutations have been identified in resistant C. albicans, C. tropicalis, P. kudriavzevii and N. glabrata, while FKS2 mutations have only been identified in N. glabrata [157,158,159,160][115][116][117][118]. No definitive intrinsic resistance has been established in any Candida species, but secondary resistance can be acquired in individual isolates through point mutations. Notably, C. parapsilosis and Meyerozyma guilliermondii have a higher rate of spontaneously occurring FKS1 point mutations and may be considered more intrinsically resistant [77,78][58][59]. Significant GOF mutations in the FKS1 gene sequence in clinical isolates have been described, particularly in the hotspot regions of 637–654 and 1345–1365 [74,157,162,163,164,165,166,167,168][56][115][119][120][121][122][123][124][125]. Walker et al. (2010) noted that non-synonymous substitutions at aa position 645 have been commonly observed. Here, serine substitutions with phenylalanine, proline or tyrosine have been cited [163,164,169][120][121][126]. Hotspot mutations are usually dominant, and C. albicans fungal cells only require one mutant allele for resistance to be conferred across the three echinocandins [74][56]. Evidence of FKS2-resistant mutations were previously limited to in vitro experiments, but recently, such mutations have been identified in clinical N. glabrata isolates [34,35,173][29][30][127]. This coincides with evidence suggesting that FKS2 has higher levels of expression in N. glabrata than FKS1 and that mutations may have a greater influence on echinocandin resistance in this species [158][116].5.2. Transcriptional Regulators of fks Genes
Transcriptional regulators upstream of fks genes can also affect echinocandin resistance. In particular, point mutations in transcription factor PDR1 have been found in numerous resistant N. glabrata isolates (Figure 3).5.3. Protein Analysis Associated with Echinocandin Resistance
Cell wall remodeling enzymes upregulated in previous protein studies include glucanosyl transferases Phr1, Phr2 and Crh, as well as chitin-glucanosyl transferase family proteins [178,179][128][129]. Proteomic analysis conducted using mass spectrometry (LC-MS/MS) revealed that levels of cell wall organization and maintenance proteins can differ between drug-resistant and susceptible strains in response to caspofungin treatment [180,181][130][131]. Differentially expressed enzymes related to cell wall synthesis and remodeling include Sun41, Gsc1, Pmt1, Mnt1, Als3, Als4, Ecm33 and Pga31 [181][131]. Validated caspofungin tolerance regulators Cas5, Mkc1, Swi4, Gin4, Stt4, Ahr1 and Pkc1 were detected in an alternate screen [182][132]. Furthermore, metabolic enzymes with immunogenic activity including Eno1, Fba1, Gpm1 and Pgk1 have also been observed to be released in Candida cells exposed to caspofungin [181,183][131][133]. Finally, the Hsp90 molecule may have a regulatory role with key resistance regulators like Mkc1 from the Pkc1 signaling pathway [184,185][134][135]. These protein subsets may be good Candidates for diagnostic markers that predict echinocandin resistance in Candida species. Further validation across a wide range of antifungal-resistant clinical isolates is still needed [181][131].6. The Nucleic Acid Biosynthesis Pathway and Antifungal Resistance
The biosynthesis of nucleic acids (DNA and RNA) and subsequent protein synthesis in pathogenic fungi can be targeted with nucleoside analogue 5-fluorocytosine (5-FC). As a prodrug, it requires activation within the fungal cell via metabolism by the pyrimidine salvage pathway [83][65]. Then, it is incorporated as a toxic substrate, and the affected nucleotides have damaging effects on cell viability [83][65]. Membrane permeases encoded by FCY2 (cytosine permease) and other homologs (FCY21 and FCY22) are responsible for the active transport of 5-FC into the cell (Figure 3) [83][65]. 5-FC is then converted to toxic 5-fluoro-uridylate by enzymes encoded by fcy1 (cytosine deaminase) and FUR1 (uracil phosphoribosyltransferase (UPRT)) [83][65]. The FCY1 homologue in C. albicans and other Candida species is the FCA1 gene [189,190][136][137]. The lack of cytosine deaminase in mammalian cells prevents 5-FC conversion and subsequent toxic effects [191][138].
Resistance to 5-FC could arise with mutation or loss of any of the three key enzymes (FCY1, FCY2 or FUR1), as discovered in model organism yeast Saccharomyces cerevisiae [192,193][139][140]. Increased pyrimidine production in the fungal cell can also serve to circumvent toxic antifungal activity [82,83][64][65]. Kern et al. (1991) were among the first to identify the correlation between a point mutation (Arg134Ser) in the FUR1 gene and 5-FC resistance in S. cerevisiae yeast cells [194][141].
7. Biofilm Formation and Antifungal Resistance
7.1. Biofilm Formation during Antifungal Treatment
Over the course of antifungal treatment, a biofilm can develop and enable yeast cells to become more resistant [209,215][142][143]. Numerous classes of antifungals, including polyenes and azoles, have been cited as less effective over time, even within 72 h of biofilm development/maturation [209][142]. For fluconazole treatment, resistance in C. albicans with biofilms can be increased by up to 1000-fold compared to planktonic cells [215,217][143][144]. Multiple features of biofilms enable antifungal resistance aside from genetic mutations that typically drive resistance in planktonic cells, which adds to the complexity of treating this type of infection [219][145]. These include physical protection, increased cell density of the microbe community, persister cells and extracellular vesicular secretion [208,219,220][145][146][147].7.2. The Roles of β-1,3 Glucan and Biofilm-Associated Antifungal Resistance
One of the main ECM components is β-1,3 glucan, which is synthesized by echinocandin target gene FKS1 [223][148]. The polysaccharide molecules are primarily responsible for the sequestration of antifungal molecules, making them a prime target for treatment of Candida infections with biofilms [224][149]. Supplemental treatment with the β-1,3 glucanase enzyme can break down this molecule and subsequently disrupt the biofilm architecture [225,226][150][151].7.3. Relevant Antifungal Resistance Genes in Biofilm-Associated Candida Infections
Candida strains associated with biofilm formation and resistance to antifungal treatment may have similar key genes implicated in planktonic resistant isolates. Changes in the expression of ergosterol biosynthesis pathway genes and in biofilm membrane composition appear to be linked to subsequent azole and polyene resistance [230,231,232,233][152][153][154][155]. Differential gene expression of beta-glucan synthesis-associated genes SKN1 and KRE1 was observed in biofilm-associated-resistant Candida exposed to amphotericin B, in agreement with previous studies [218,230][152][156]. These relevant genes may be highlighted in the search for a suitable resistance biomarker. Drug efflux pumps CDR1, CDR2 and MDR1, which are associated with azole resistance, may also be upregulated in biofilm-associated Candida strains [235,236,237,238,239,240][157][158][159][160][161][162]. Interestingly, neither polyenes nor echinocandins have been implicated as a substrate for drug efflux pumps in Candida infections of this type. This suggests that echinocandins could be a preferable treatment choice over azoles [236,237,241][158][159][163].8. Summary
With antifungal resistance being a continued problem, there is a need for the development of quick and reliable molecular diagnostic tests that detect organisms with intrinsic and/or secondary resistance due to the genetic mechanisms [34][29]. Additionally, faster methods of species identification would be useful, given the differences in frequency and impact of antifungal resistance among Candida species. Currently, PCR-based methods using fungal cultures are still the first option for species identification and detection of antifungal resistance in individual strains [105,167][124][164]. The usefulness of real-time testing for resistance in Candida species to modify the treatment course has been well documented.
Resistance can be acquired through a dynamic combination of numerous point mutations and other genetic or transcriptional alterations. A large-scale comparison between matched fluconazole-resistant and -susceptible C. albicans clinical isolates using microarray analysis identified almost two hundred genes (n = 198) that were differentially expressed [248][165]. In resistant isolates, multidrug resistance and oxidative stress response genes, among others, were found to be upregulated compared to susceptible samples [248][165]. This highlights the fact that a dynamic response that leads to antifungal resistance and identification of reliable biomarkers or gene expression profiles that different strains have in common would be beneficial to improve future treatment decisions.
To develop a dependable point-of-care (POCT) diagnostic assay for antifungal resistance, reliable biomarkers in Candida species need to be established. It is important to note that different non-synonymous substitutions and other genetic alterations may result in similar genetic effects.References
- Bhattacharya, S.; Sae-Tia, S.; Fries, B.C. Candidiasis and Mechanisms of Antifungal Resistance. Antibiotics 2020, 9, 312.
- Vázquez-González, D.; Perusquía-Ortiz, A.M.; Hundeiker, M.; Bonifaz, A. Opportunistic Yeast Infections: Candidiasis, Cryptococcosis, Trichosporonosis and Geotrichosis. JDDG J. Ger. Soc. Dermatol. 2013, 11, 381–394.
- Fisher, M.C.; Gow, N.A.R.; Gurr, S.J. Tackling Emerging Fungal Threats to Animal Health, Food Security and Ecosystem Resilience. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20160332.
- Kourkoumpetis, T.K.; Velmahos, G.C.; Ziakas, P.D.; Tampakakis, E.; Manolakaki, D.; Coleman, J.J.; Mylonakis, E. The Effect of Cumulative Length of Hospital Stay on the Antifungal Resistance of Candida Strains Isolated from Critically Ill Surgical Patients. Mycopathologia 2011, 171, 85–91.
- Dąbrowska, M.; Sienkiewicz, M.; Kwiatkowski, P.; Dąbrowski, M. Diagnosis and Treatment of Mucosa Candida spp. Infections—A Review Article. Ann. Univ. Mariae Curie Sklodowska Sect. C Biol. 2019, 73, 61–68.
- Gonsalves, W.C.; Wrightson, A.S.; Henry, R.G. Common Oral Conditions in Older Persons. Am. Fam. Physician 2008, 78, 845–852.
- Lalla, R.V.; Latortue, M.C.; Hong, C.H.; Ariyawardana, A.; D’Amato-Palumbo, S.; Fischer, D.J.; Martof, A.; Nicolatou-Galitis, O.; Patton, L.L.; Elting, L.S.; et al. A Systematic Review of Oral Fungal Infections in Patients Receiving Cancer Therapy. Support. Care Cancer 2010, 18, 985–992.
- Rohr, Y.; Adams, J.; Young, L. Oral Discomfort in Palliative Care: Results of an Exploratory Study of the Experiences of Terminally Ill Patients. Int. J. Palliat. Nurs. 2010, 16, 439–444.
- Pfaller, M.; Neofytos, D.; Diekema, D.; Azie, N.; Meier-Kriesche, H.U.; Quan, S.P.; Horn, D. Epidemiology and Outcomes of Candidemia in 3648 Patients: Data from the Prospective Antifungal Therapy (PATH Alliance®) Registry, 2004–2008. Diagn. Microbiol. Infect. Dis. 2012, 74, 323–331.
- Fisher, M.C.; Alastruey-Izquierdo, A.; Berman, J.; Bicanic, T.; Bignell, E.M.; Bowyer, P.; Bromley, M.; Brüggemann, R.; Garber, G.; Cornely, O.A.; et al. Tackling the Emerging Threat of Antifungal Resistance to Human Health. Nat. Rev. Microbiol. 2022, 20, 557–571.
- Kumar, R.; Srivastava, V. Application of Anti-Fungal Vaccines as a Tool against Emerging Anti-Fungal Resistance. Front. Fungal Biol. 2023, 4, 1241539.
- Benedict, K.; Jackson, B.R.; Chiller, T.; Beer, K.D. Estimation of Direct Healthcare Costs of Fungal Diseases in the United States. Clin. Infect. Dis. 2019, 68, 1791–1797.
- Gabaldón, T.; Naranjo-Ortíz, M.A.; Marcet-Houben, M. Evolutionary Genomics of Yeast Pathogens in the Saccharomycotina. FEMS Yeast Res. 2016, 16, fow064.
- Defosse, T.A.; Le Govic, Y.; Courdavault, V.; Clastre, M.; Vandeputte, P.; Chabasse, D.; Bouchara, J.P.; Giglioli-Guivarc’h, N.; Papon, N. Yeasts from the CTG Clade (Candida Clade): Biology, Impact in Human Health, and Biotechnological Applications. J. Mycol. Med. 2018, 28, 257–268.
- Santos, M.A.S.; Gomes, A.C.; Santos, M.C.; Carreto, L.C.; Moura, G.R. The Genetic Code of the Fungal CTG Clade. C R. Biol. 2011, 334, 607–611.
- Borman, A.M.; Johnson, E.M. Name Changes for Fungi of Medical Importance, 2018 to 2019. J. Clin. Microbiol. 2021, 59.
- Fidel, P.L.; Vazquez, J.A.; Sobel, J.D. Candida Glabrata: Review of Epidemiology, Pathogenesis, and Clinical Disease with Comparison to C. Albicans. Clin. Microbiol. Rev. 1999, 12, 80–96.
- Healey, K.R.; Ortigosa, C.J.; Shor, E.; Perlin, D.S. Genetic Drivers of Multidrug Resistance in Candida Glabrata. Front. Microbiol. 2016, 7, 1995.
- World Health Organization. WHO Fungal Priority Pathogens List to Guide Research, Development and Public Health Action; World Health Organization: Geneva, Switzerland, 2022; Volume 1.
- CDC Antibiotic Resistance Threats in the United States; U.S. Department of Health and Human Services: Atlanta, GA, USA, 2019.
- Brandt, M.E.; Lockhart, S.R. Recent Taxonomic Developments with Candida and Other Opportunistic Yeasts. Curr. Fungal Infect. Rep. 2012, 6, 170–177.
- Pfaller, M.A.; Diekema, D.J.; Gibbs, D.L.; Newell, V.A.; Ellis, D.; Tullio, V.; Rodloff, A.; Fu, W.; Ling, T.A. Results from the Artemis Disk Global Antifungal Surveillance Study, 1997 to 2007: A 10.5-Year Analysis of Susceptibilities of Candida Species to Fluconazole and Voriconazole as Determined by CLSI Standardized Disk Diffusion. J. Clin. Microbiol. 2010, 48, 1366–1377.
- Walsh, T.J.; Groll, A.; Hiemenz, J.; Fleming, R.; Roilides, E.; Anaissie, E. Infections Due to Emerging and Uncommon Medically Important Fungal Pathogens. Clin. Microbiol. Infect. 2004, 10, 48–66.
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous Emergence of Multidrug-Resistant Candida Auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin. Infect. Dis. 2017, 64, 134–140.
- Infection Prevention and Control. Candida auris. Public Health Ontario. Available online: https://www.publichealthontario.ca/en/Diseases-and-Conditions/Health-Care-Associated-Infections/Candida-auris (accessed on 10 October 2023).
- Centers for Disease Control and Prevention. Antifungal Susceptibility Testing and Interpretation. Available online: https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html#print (accessed on 10 October 2023).
- CLSI. CLSI Performance Standards for Antifungal Susceptibility Testing of Yeasts, 3rd ed.; CLSI Supplement M27M44S; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022; Volume 40.
- Wiederhold, N.P. Antifungal Susceptibility Testing: A Primer for Clinicians. Open Forum Infect. Dis. 2021, 8, ofab444.
- Garcia-Effron, G. Molecular Markers of Antifungal Resistance: Potential Uses in Routine Practice and Future Perspectives. J. Fungi 2021, 7, 197.
- Castanheira, M.; Deshpande, L.M.; Davis, A.P.; Carvalhaes, C.G.; Pfaller, M.A. Azole Resistance in Candida Glabrata Clinical Isolates from Global Surveillance Is Associated with Efflux Overexpression. J. Glob. Antimicrob. Resist. 2022, 29, 371–377.
- White, T.C.; Holleman, S.; Dy, F.; Mirels, L.F.; Stevens, D.A. Resistance Mechanisms in Clinical Isolates of Candida Albicans. Antimicrob. Agents Chemother. 2002, 46, 1704–1713.
- Shields, R.K.; Nguyen, M.H.; Press, E.G.; Kwa, A.L.; Cheng, S.; Du, C.; Clancy, C.J. The Presence of an FKS Mutation Rather than MIC Is an Independent Risk Factor for Failure of Echinocandin Therapy among Patients with Invasive Candidiasis Due to Candida Glabrata. Antimicrob. Agents Chemother. 2012, 56, 4862–4869.
- Bienvenu, A.L.; Leboucher, G.; Picot, S. Comparison of Fks Gene Mutations and Minimum Inhibitory Concentrations for the Detection of Candida Glabrata Resistance to Micafungin: A Systematic Review and Meta-Analysis. Mycoses 2019, 62, 835–846.
- Pfaller, M.A.; Andes, D.; Diekema, D.J.; Espinel-Ingroff, A.; Sheehan, D. Wild-Type MIC Distributions, Epidemiological Cutoff Values and Species-Specific Clinical Breakpoints for Fluconazole and Candida: Time for Harmonization of CLSI and EUCAST Broth Microdilution Methods. Drug Resist. Updates 2010, 13, 180–195.
- Pfaller, M.A.; Andes, D.; Arendrup, M.C.; Diekema, D.J.; Espinel-Ingroff, A.; Alexander, B.D.; Brown, S.D.; Chaturvedi, V.; Fowler, C.L.; Ghannoum, M.A.; et al. Clinical Breakpoints for Voriconazole and Candida Spp. Revisited: Review of Microbiologic, Molecular, Pharmacodynamic, and Clinical Data as They Pertain to the Development of Species-Specific Interpretive Criteria. Diagn. Microbiol. Infect. Dis. 2011, 70, 330–343.
- Pfaller, M.A.; Diekema, D.J.; Andes, D.; Arendrup, M.C.; Brown, S.D.; Lockhart, S.R.; Motyl, M.; Perlin, D.S. Clinical Breakpoints for the Echinocandins and Candida Revisited: Integration of Molecular, Clinical, and Microbiological Data to Arrive at Species-Specific Interpretive Criteria. Drug Resist. Updates 2011, 14, 164–176.
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 13.0. 2023. Available online: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_5.0_Breakpoint_Table_01.pdf (accessed on 10 October 2023).
- Maria, S.; Barnwal, G.; Kumar, A.; Mohan, K.; Vinod, V.; Varghese, A.; Biswas, R. Species Distribution and Antifungal Susceptibility among Clinical Isolates of Candida Parapsilosis Complex from India. Rev. Iberoam. Micol. 2018, 35, 147–150.
- Borman, A.M.; Muller, J.; Walsh-Quantick, J.; Szekely, A.; Patterson, Z.; Palmer, M.D.; Fraser, M.; Johnson, E.M. Fluconazole Resistance in Isolates of Uncommon Pathogenic Yeast Species from the United Kingdom. Antimicrob. Agents Chemother. 2019, 63, e00211-19.
- Vigezzi, C.; Icely, P.A.; Dudiuk, C.; Rodríguez, E.; Miró, M.S.; Castillo, G.D.V.; Azcurra, A.I.; Abiega, C.; Caeiro, J.P.; Riera, F.O.; et al. Frequency, Virulence Factors and Antifungal Susceptibility of Candida Parapsilosis Species Complex Isolated from Patients with Candidemia in the Central Region of Argentina. J. Mycol. Med. 2019, 29, 285–291.
- CLSI. Epidemiological Cutoff Values for Antifungal Susceptibility Testing, 2nd ed.; CLSI Supplement M59; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020.
- Pfaller, M.A.; Diekema, D.J.; Turnidge, J.D.; Castanheira, M.; Jones, R.N. Twenty Years of the SENTRY Antifungal Surveillance Program: Results for Candida Species from 1997-2016. Open Forum Infect. Dis. 2019, 6, S79–S94.
- de Tilly, A.N.; Tharmalingam, S. Review of Treatments for Oropharyngeal Fungal Infections in HIV/AIDS Patients. Microbiol. Res. 2022, 13, 219–234.
- Shalini, K.; Kumar, N.; Drabu, S.; Sharma, P.K. Advances in Synthetic Approach to and Antifungal Activity of Triazoles. Beilstein J. Org. Chem. 2011, 7, 668–677.
- Robbins, N.; Caplan, T.; Cowen, L.E. Molecular Evolution of Antifungal Drug Resistance. Annu. Rev. Microbiol. 2017, 71, 753–775.
- Robbins, N.; Wright, G.D.; Cowen, L.E. Antifungal Drugs: The Current Armamentarium and Development of New Agents. Microbiol. Spectr. 2016, 4, 903–922.
- Sanglard, D.; Coste, A.; Ferrari, S. Antifungal Drug Resistance Mechanisms in Fungal Pathogens from the Perspective of Transcriptional Gene Regulation. FEMS Yeast Res. 2009, 9, 1029–1050.
- Azanza, J.R.; García-Quetglas, E.; Sádaba, B. Pharmacology of Azoles. Rev. Iberoam. Micol. 2007, 24, 223–227.
- Efimova, S.S.; Schagina, L.V.; Ostroumova, O.S. Investigation of Channel-Forming Activity of Polyene Macrolide Antibiotics in Planar Lipid Bilayers in the Presence of Dipole Modifiers. Acta Naturae 2014, 6, 67–79.
- Kermani, F.; Taghizadeh-Armaki, M.; Hosseini, S.A.; Amirrajab, N.; Javidnia, J.; Zaghrami, M.F.; Shokohi, T. Antifungal Resistance of Clinical Candida Albicans Isolates in Iran: A Systematic Review and Meta-Analysis. Iran. J. Public Health 2023, 52, 290–305.
- Birch, M.; Sibley, G. Antifungal Chemistry Review. In Comprehensive Medicinal Chemistry III; Elsevier: Amsterdam, The Netherlands, 2017.
- Waller, D.G.; Sampson, A.P. Chemotherapy of Infections. In Medical Pharmacology and Therapeutics; Elsevier: Amsterdam, The Netherlands, 2018.
- Espinel-Ingroff, A.; Arendrup, M.; Canton, E.; Cordob, S.; Dannaoui, E.; Garcia-Rodriguez, J.; Gonzalez, G.M.; Govender, N.P.; Martin-Mazuelos, E.; Lackner, M.; et al. Multicenter Study of Method-Dependent Epidemiological Cutoff Values for Detection of Resistance in Candida Spp. and Aspergillus Spp. to Amphotericin B and Echinocandins for the Etest Agar Diffusion Method. Antimicrob. Agents Chemother. 2017, 61, e01792-16.
- Kanafani, Z.A.; Perfect, J.R. Resistance to Antifungal Agents: Mechanisms and Clinical Impact. Clin. Infect. Dis. 2008, 46, 120–128.
- Popolo, L.; Gualtieri, T.; Ragni, E. The Yeast Cell-Wall Salvage Pathway. Med. Mycol. Suppl. 2001, 39, 111–121.
- Walker, L.A.; Gow, N.A.R.; Munro, C.A. Fungal Echinocandin Resistance. Fungal Genet. Biol. 2010, 47, 117–126.
- Douglas, C.M.; D’Ippolito, J.A.; Shei, G.J.; Meinz, M.; Onishi, J.; Marrinan, J.A.; Li, W.; Abruzzo, G.K.; Flattery, A.; Bartizal, K.; et al. Identification of the FKS1 Gene of Candida Albicans as the Essential Target of 1,3-β-D-Glucan Synthase Inhibitors. Antimicrob. Agents Chemother. 1997, 41, 2471–2479.
- Barchiesi, F.; Spreghini, E.; Tomassetti, S.; Della Vittoria, A.; Arzeni, D.; Manso, E.; Scalise, G. Effects of Caspofungin against Candida Guilliermondii and Candida Parapsilosis. Antimicrob. Agents Chemother. 2006, 50, 2719–2727.
- Cantón, E.; Pemán, J.; Sastre, M.; Romero, M.; Espinel-Ingroff, A. Killing Kinetics of Caspofungin, Micafungin, and Amphotericin B against Candida Guilliermondii. Antimicrob. Agents Chemother. 2006, 50, 2829–2832.
- Arastehfar, A.; Lass-Flörl, C.; Garcia-Rubio, R.; Daneshnia, F.; Ilkit, M.; Boekhout, T.; Gabaldon, T.; Perlin, D.S. The Quiet and Underappreciated Rise of Drug-Resistant Invasive Fungal Pathogens. J. Fungi 2020, 6, 138.
- Bohner, F.; Papp, C.; Gácser, A. The Effect of Antifungal Resistance Development on the Virulence of Candida Species. FEMS Yeast Res. 2022, 22, foac019.
- Vermes, A.; Guchelaar, H.J.; Dankert, J. Flucytosine: A Review of Its Pharmacology, Clinical Indications, Pharmacokinetics, Toxicity and Drug Interactions. J. Antimicrob. Chemother. 2000, 46, 171–179.
- Mourad, A.; Perfect, J.R. Present and Future Therapy of Cryptococcus Infections. J. Fungi 2018, 4, 79.
- Perfect, J.R.; Dismukes, W.E.; Dromer, F.; Goldman, D.L.; Graybill, J.R.; Hamill, R.J.; Harrison, T.S.; Larsen, R.A.; Lortholary, O.; Nguyen, M.H.; et al. Clinical Practice Guidelines for the Management of Cryptococcal Disease: 2010 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2010, 50, 291–322.
- Delma, F.Z.; Al-Hatmi, A.M.S.; Brüggemann, R.J.M.; Melchers, W.J.G.; de Hoog, S.; Verweij, P.E.; Buil, J.B. Molecular Mechanisms of 5-Fluorocytosine Resistance in Yeasts and Filamentous Fungi. J. Fungi 2021, 7, 909.
- Bhattacharya, S.; Esquivel, B.D.; White, T.C. Overexpression or Deletion of Ergosterol Biosynthesis Genes Alters Doubling Time, Response to Stress Agents, and Drug Susceptibility in Saccharomyces Cerevisiae. mBio 2018, 9, e01291-18.
- Xiang, M.J.; Liu, J.Y.; Ni, P.H.; Wang, S.; Shi, C.; Wei, B.; Ni, Y.X.; Ge, H.L. Erg11 Mutations Associated with Azole Resistance in Clinical Isolates of Candida Albicans. FEMS Yeast Res. 2013, 13, 386–393.
- Manastir, L.; Ergon, M.C.; Yücesoy, M. Investigation of Mutations in Erg11 Gene of Fluconazole Resistant Candida Albicans Isolates from Turkish Hospitals. Mycoses 2011, 54, 99–104.
- Morio, F.; Loge, C.; Besse, B.; Hennequin, C.; Le Pape, P. Screening for Amino Acid Substitutions in the Candida Albicans Erg11 Protein of Azole-Susceptible and Azole-Resistant Clinical Isolates: New Substitutions and a Review of the Literature. Diagn. Microbiol. Infect. Dis. 2010, 66, 373–384.
- Feng, L.J.; Wan, Z.; Wang, X.H.; Li, R.Y.; Liu, W. Relationship between Antifungal Resistance of Fluconazole Resistant Candida Albicans and Mutations in ERG11 Gene. Chin. Med. J. 2010, 123, 544–548.
- dos Santos Silva, D.B.; Carbonera Rodrigues, L.M.; De Almeida, A.A.; de Oliveira, K.M.P.; Grisolia, A.B. Novel Point Mutations in the ERG11 Gene in Clinical Isolates of Azole Resistant Candida Species. Mem. Inst. Oswaldo Cruz 2016, 111, 192–199.
- Sanglard, D.; Ischer, F.; Calabrese, D.; Micheli, M.d.; Bille, J. Multiple Resistance Mechanisms to Azole Antifungals in Yeast Clinical Isolates. Drug Resist. Updates 1998, 1, 255–265.
- Sanglard, D.; Ischer, F.; Koymans, L.; Bille, J. Amino Acid Substitutions in the Cytochrome P-450 Lanosterol 14α- Demethylase (CYP51A1) from Azole-Resistant Candida Albicans Clinical Isolates Contribute to Resistance to Azole Antifungal Agents. Antimicrob. Agents Chemother. 1998, 42, 241–253.
- Li, X.; Brown, N.; Chau, A.S.; López-Ribot, J.L.; Ruesga, M.T.; Quindos, G.; Mendrick, C.A.; Hare, R.S.; Loebenberg, D.; DiDomenico, B.; et al. Changes in Susceptibility to Posaconazole in Clinical Isolates of Candida Albicans. J. Antimicrob. Chemother. 2004, 53, 74–80.
- Dunkel, N.; Liu, T.T.; Barker, K.S.; Homayouni, R.; Morschhäuser, J.; Rogers, P.D. A Gain-of-Function Mutation in the Transcription Factor Upc2p Causes Upregulation of Ergosterol Biosynthesis Genes and Increased Fluconazole Resistance in a Clinical Candida Albicans Isolate. Eukaryot. Cell 2008, 7, 1180–1190.
- Hoot, S.J.; Smith, A.R.; Brown, R.P.; White, T.C. An A643V Amino Acid Substitution in Upc2p Contributes to Azole Resistance in Well-Characterized Clinical Isolates of Candida Albicans. Antimicrob. Agents Chemother. 2011, 55, 940–942.
- Znaidi, S.; Weber, S.; Al-Abdin, O.Z.; Bomme, P.; Saidane, S.; Drouin, S.; Lemieux, S.; De Deken, X.; Robert, F.; Raymond, M. Genomewide Location Analysis of Candida Albicans Upc2p, a Regulator of Sterol Metabolism and Azole Drug Resistance. Eukaryot. Cell 2008, 7, 836–847.
- Barker, K.S.; Crisp, S.; Wiederhold, N.; Lewis, R.E.; Bareither, B.; Eckstein, J.; Barbuch, R.; Bard, M.; Rogers, P.D. Genome-Wide Expression Profiling Reveals Genes Associated with Amphotericin B and Fluconazole Resistance in Experimentally Induced Antifungal Resistant Isolates of Candida Albicans. J. Antimicrob. Chemother. 2004, 54, 376–385.
- Kelly, S.L.; Lamb, D.C.; Corran, A.J.; Baldwin, B.C.; Kelly, D.E. Mode of Action and Resistance to Azole Antifungals Associated with the Formation of 14α-Methylergosta-8,24(28)-Dien-3β,6α-Diol. Biochem. Biophys. Res. Commun. 1995, 207, 910–915.
- Watson, P.F.; Rose, M.E.; Ellis, S.W.; England, H.; Kelly, S.L. Defective Sterol C5-6 Desaturation and Azole Resistance: A New Hypothesis for the Mode of Action of Azole Antifungals. Biochem. Biophys. Res. Commun. 1989, 164, 1170–1175.
- Sanglard, D.; Ischer, F.; Parkinson, T.; Falconer, D.; Bille, J. Candida Albicans Mutations in the Ergosterol Biosynthetic Pathway and Resistance to Several Antifungal Agents. Antimicrob. Agents Chemother. 2003, 47, 2404–2412.
- Whaley, S.G.; Berkow, E.L.; Rybak, J.M.; Nishimoto, A.T.; Barker, K.S.; Rogers, P.D. Azole Antifungal Resistance in Candida Albicans and Emerging Non-Albicans Candida Species. Front. Microbiol. 2017, 7, 2173.
- Yoo, J.I.; Choi, C.W.; Lee, K.M.; Lee, Y.S. Gene Expression and Identification Related to Fluconazole Resistance of Candida Glabrata Strains. Osong Public. Health Res. Perspect. 2010, 1, 36–41.
- Anderson, J.B.; Sirjusingh, C.; Parsons, A.B.; Boone, C.; Wickens, C.; Cowen, L.E.; Kohn, L.M. Mode of Selection and Experimental Evolution of Antifungal Drug Resistance in Saccharomyces Cerevisiae. Genetics 2003, 163, 1287–1298.
- Xu, D.; Jiang, B.; Ketela, T.; Lemieux, S.; Veillette, K.; Martel, N.; Davison, J.; Sillaots, S.; Trosok, S.; Bachewich, C.; et al. Genome-Wide Fitness Test and Mechanism-of-Action Studies of Inhibitory Compounds in Candida Albicans. PLoS Pathog. 2007, 3, e92.
- Jia, Y.; Tang, R.J.; Wang, L.; Zhang, X.; Wang, Y.; Jia, X.M.; Jiang, Y.Y. Calcium-Activated-Calcineurin Reduces the In Vitro and In Vivo Sensitivity of Fluconazole to Candida Albicans via Rta2p. PLoS ONE 2012, 7, e48369.
- Perea, S.; López-Ribot, J.L.; Kirkpatrick, W.R.; McAtee, R.K.; Santillán, R.A.; Martínez, M.; Calabrese, D.; Sanglard, D.; Patterson, T.F. Prevalence of Molecular Mechanisms of Resistance to Azole Antifungal Agents in Candida Albicans Strains Displaying High-Level Fluconazole Resistance Isolated from Human Immunodeficiency Virus-Infected Patients. Antimicrob. Agents Chemother. 2001, 45, 2676–2684.
- Prasad, R.; Nair, R.; Banerjee, A. Multidrug Transporters of Candida Species in Clinical Azole Resistance. Fungal Genet. Biol. 2019, 132, 103252.
- Sanglard, D.; Ischer, F.; Monod, M.; Bille, J. Susceptibilities of Candida Albicans Multidrug Transporter Mutants to Various Antifungal Agents and Other Metabolic Inhibitors. Antimicrob. Agents Chemother. 1996, 40, 2300–2305.
- Khandelwal, N.K.; Wasi, M.; Nair, R.; Gupta, M.; Kumar, M.; Mondal, A.K.; Gaur, N.A.; Prasad, R. Vacuolar Sequestration of Azoles, a Novel Strategy of Azole Antifungal Resistance Conserved across Pathogenic and Nonpathogenic Yeast. Antimicrob. Agents Chemother. 2019, 63, e01347-18.
- Bencova, A.; Goffa, E.; Morvova, M.; Valachovic, M.; Griač, P.; Toth Hervay, N.; Gbelska, Y. The Absence of PDR16 Gene Restricts the Overexpression of CaSNQ2 Gene in the Presence of Fluconazole in Candida Albicans. Mycopathologia 2020, 185, 455–465.
- Znaidi, S.; De Deken, X.; Weber, S.; Rigby, T.; Nantel, A.; Raymond, M. The Zinc Cluster Transcription Factor Tac1p Regulates PDR16 Expression in Candida Albicans. Mol. Microbiol. 2007, 66, 440–452.
- Perepnikhatka, V.; Fischer, F.J.; Niimi, M.; Baker, R.A.; Cannon, R.D.; Wang, Y.K.; Sherman, F.; Rustchenko, E. Specific Chromosome Alterations in Fluconazole-Resistant Mutants of Candida Albicans. J. Bacteriol. 1999, 181, 4041–4049.
- Sanguinetti, M.; Posteraro, B.; Fiori, B.; Ranno, S.; Torelli, R.; Fadda, G. Mechanisms of Azole Resistance in Clinical Isolates of Candida Glabrata Collected during a Hospital Survey of Antifungal Resistance. Antimicrob. Agents Chemother. 2005, 49, 668–679.
- Miyazaki, H.; Miyazaki, Y.; Geber, A.; Parkinson, T.; Hitchcock, C.; Falconer, D.J.; Ward, D.J.; Marsden, K.; Bennett, J.E. Fluconazole Resistance Associated with Drug Efflux and Increased Transcription of a Drug Transporter Gene, PDH1, in Candida Glabrata. Antimicrob. Agents Chemother. 1998, 42, 1695–1701.
- Whaley, S.G.; Zhang, Q.; Caudle, K.E.; Rogers, P.D. Relative Contribution of the ABC Transporters Cdr1, Pdh1, and Snq2 to Azole Resistance in Candida Glabrata. Antimicrob. Agents Chemother. 2018, 62, e01070-18.
- Costa, C.; Dias, P.J.; Sá-Correia, I.; Teixeira, M.C. MFS Multidrug Transporters in Pathogenic Fungi: Do They Have Real Clinical Impact? Front. Physiol. 2014, 5, 197.
- Costa, C.; Ribeiro, J.; Miranda, I.M.; Silva-Dias, A.; Cavalheiro, M.; Costa-de-Oliveira, S.; Rodrigues, A.G.; Teixeira, M.C. Clotrimazole Drug Resistance in Candida Glabrata Clinical Isolates Correlates with Increased Expression of the Drug: H+ Antiporters CgAqr1, CgTpo1_1, CgTpo3, and CgQdr2. Front. Microbiol. 2016, 7, 526.
- Bhattacharya, S.; Friesa, B.C. Enhanced Efflux Pump Activity in Old Candida Glabrata Cells. Antimicrob. Agents Chemother. 2018, 62, e02227-17.
- Culakova, H.; Dzugasova, V.; Valencikova, R.; Gbelska, Y.; Subik, J. Stress Response and Expression of Fluconazole Resistance Associated Genes in the Pathogenic Yeast Candida Glabrata Deleted in the CgPDR16 Gene. Microbiol. Res. 2015, 174, 17–23.
- Coste, A.; Turner, V.; Ischer, F.; Morschhäuser, J.; Forche, A.; Selmecki, A.; Berman, J.; Bille, J.; Sanglard, D. A Mutation in Tac1p, a Transcription Factor Regulating CDR1 and CDR2, Is Coupled with Loss of Heterozygosity at Chromosome 5 to Mediate Antifungal Resistance in Candida Albicans. Genetics 2006, 172, 2139–2156.
- Rybak, J.M.; Muñoz, J.F.; Barker, K.S.; Parker, J.E.; Esquivel, B.D.; Berkow, E.L.; Lockhart, S.R.; Gade, L.; Palmer, G.E.; White, T.C.; et al. Mutations in TAC1B: A Novel Genetic Determinant of Clinical Fluconazole Resistance in Candida Auris. mBio 2020, 11, e00365-20.
- Kalkandelen, K.T.; Doluca Dereli, M. Investigation of Mutations in Transcription Factors of Efflux Pump Genes in Fluconazole-Resistant Candida Albicans Strains Overexpressing the Efflux Pumps. Mikrobiyol. Bul. 2015, 49, 609–618.
- Schubert, S.; Rogers, P.D.; Morschhäuser, J. Gain-of-Function Mutations in the Transcription Factor MRR1 Are Responsible for Overexpression of the MDR1 Efflux Pump in Fluconazole-Resistant Candida Dubliniensis Strains. Antimicrob. Agents Chemother. 2008, 52, 4274–4280.
- Moran, G.P.; Sullivan, D.J.; Henman, M.C.; McCreary, C.E.; Harrington, B.J.; Shanley, D.B.; Coleman, D.C. Antifungal Drug Susceptibilities of Oral Candida Dubliniensis Isolates from Human Immunodeficiency Virus (HIV)-Infected and Non-HIV-Infected Subjects and Generation of Stable Fluconazole-Resistant Derivatives in Vitro. Antimicrob. Agents Chemother. 1997, 41, 617–623.
- Moran, G.P.; Sanglard, D.; Donnelly, S.M.; Shanley, D.B.; Sullivan, D.J.; Coleman, D.C. Identification and Expression of Multidrug Transporters Responsible for Fluconazole Resistance in Candida Dubliniensis. Antimicrob. Agents Chemother. 1998, 42, 1819–1830.
- Morschhäuser, J.; Barker, K.S.; Liu, T.T.; Blaß-Warmuth, J.; Homayouni, R.; Rogers, P.D. The Transcription Factor Mrr1p Controls Expression of the MDR1 Efflux Pump and Mediates Multidrug Resistance in Candida Albicans. PLoS Pathog. 2007, 3, e164.
- Schubert, S.; Barker, K.S.; Znaidi, S.; Schneider, S.; Dierolf, F.; Dunkel, N.; Aïd, M.; Boucher, G.; Rogers, P.D.; Raymond, M.; et al. Regulation of Efflux Pump Expression and Drug Resistance by the Transcription Factors Mrr1, Upc2, and Cap1 in Candida Albicans. Antimicrob. Agents Chemother. 2011, 55, 2212–2223.
- Dunkel, N.; Blaß, J.; Rogers, P.D.; Morschhäuser, J. Mutations in the Multi-Drug Resistance Regulator MRR1, Followed by Loss of Heterozygosity, Are the Main Cause of MDR1 Overexpression in Fluconazole-Resistant Candida Albicans Strains. Mol. Microbiol. 2008, 69, 827–840.
- Shukla, S.; Yadav, V.; Mukhopadhyay, G.; Prasad, R. Ncb2 Is Involved in Activated Transcription of CDR1 in Azole-Resistant Clinical Isolates of Candida Albicans ∇. Eukaryot. Cell 2011, 10, 1357–1366.
- Mogavero, S.; Tavanti, A.; Senesi, S.; Rogers, P.D.; Morschhäuser, J. Differential Requirement of the Transcription Factor Mcm1 for Activation of the Candida Albicans Multidrug Efflux Pump MDR1 by Its Regulators Mrr1 and Cap1. Antimicrob. Agents Chemother. 2011, 55, 2061–2066.
- Alarco, A.M.; Raymond, M. The BZip Transcription Factor Cap1p Is Involved in Multidrug Resistance and Oxidative Stress Response in Candida Albicans. J. Bacteriol. 1999, 181, 700–708.
- Thomas, E.; Roman, E.; Claypool, S.; Manzoor, N.; Pla, J.; Panwar, S.L. Mitochondria Influence CDR1 Efflux Pump Activity, Hog1-Mediated Oxidative Stress Pathway, Iron Homeostasis, and Ergosterol Levels in Candida Albicans. Antimicrob. Agents Chemother. 2013, 57, 5580–5599.
- Alexander, B.D.; Johnson, M.D.; Pfeiffer, C.D.; Jiménez-Ortigosa, C.; Catania, J.; Booker, R.; Castanheira, M.; Messer, S.A.; Perlin, D.S.; Pfaller, M.A. Increasing Echinocandin Resistance in Candida Glabrata: Clinical Failure Correlates with Presence of FKS Mutations and Elevated Minimum Inhibitory Concentrations. Clin. Infect. Dis. 2013, 56, 1724–1732.
- Cleary, J.D.; Garcia-Effron, G.; Chapman, S.W.; Perlin, D.S. Reduced Candida Glabrata Susceptibility Secondary to an FKS1 Mutation Developed during Candidemia Treatment. Antimicrob. Agents Chemother. 2008, 52, 2263–2265.
- Garcia-Effron, G.; Lee, S.; Park, S.; Cleary, J.D.; Perlin, D.S. Effect of Candida Glabrata FKS1 and FKS2 Mutations on Echinocandin Sensitivity and Kinetics of 1,3-β-D-Glucan Synthase: Implication for the Existing Susceptibility Breakpoint. Antimicrob. Agents Chemother. 2009, 53, 3690–3699.
- Garcia-Effron, G.; Chua, D.J.; Tomada, J.R.; DiPersio, J.; Perlin, D.S.; Ghannoum, M.; Bonilla, H. Novel FKS Mutations Associated with Echinocandin Resistance in Candida Species. Antimicrob. Agents Chemother. 2010, 54, 2225–2227.
- Thompson, G.R.; Wiederhold, N.P.; Vallor, A.C.; Villareal, N.C.; Lewis, J.S.; Patterson, T.F. Development of Caspofungin Resistance Following Prolonged Therapy for Invasive Candidiasis Secondary to Candida Glabrata Infection. Antimicrob. Agents Chemother. 2008, 52, 3783–3785.
- Munro, C.A. Fungal Echinocandin Resistance. F1000 Biol. Rep. 2010, 2, 117–126.
- Park, S.; Kelly, R.; Kahn, J.N.; Robles, J.; Hsu, M.J.; Register, E.; Li, W.; Vyas, V.; Fan, H.; Abruzzo, G.; et al. Specific Substitutions in the Echinocandin Target Fks1p Account for Reduced Susceptibility of Rare Laboratory and Clinical Candida sp. Isolates. Antimicrob. Agents Chemother. 2005, 49, 3264–3273.
- Perlin, D.S. Resistance to Echinocandin-Class Antifungal Drugs. Drug Resist. Updates 2007, 10, 121–130.
- Hakki, M.; Staab, J.F.; Marr, K.A. Emergence of a Candida Krusei Isolate with Reduced Susceptibility to Caspofungin during Therapy. Antimicrob. Agents Chemother. 2006, 50, 2522–2524.
- Kahn, J.N.; Garcia-Effron, G.; Hsu, M.J.; Park, S.; Marr, K.A.; Perlin, D.S. Acquired Echinocandin Resistance in a Candida Krusei Isolate Due to Modification of Glucan Synthase. Antimicrob. Agents Chemother. 2007, 51, 1876–1878.
- Garcia-Effron, G.; Park, S.; Perlin, D.S. Correlating Echinocandin MIC and Kinetic Inhibition of Fks1 Mutant Glucan Synthases for Candida Albicans: Implications for Interpretive Breakpoints. Antimicrob. Agents Chemother. 2009, 53, 112–122.
- Sharma, D.; Paul, R.A.; Rudramurthy, S.M.; Kashyap, N.; Bhattacharya, S.; Soman, R.; Shankarnarayan, S.A.; Chavan, D.; Singh, S.; Das, P.; et al. Impact of FKS1 Genotype on Echinocandin In Vitro Susceptibility in Candida Auris and In Vivo Response in a Murine Model of Infection. Antimicrob. Agents Chemother. 2022, 66, e0165221.
- Balashov, S.V.; Park, S.; Perlin, D.S. Assessing Resistance to the Echinocandin Antifungal Drug Caspofungin in Candida Albicans by Profiling Mutations in FKS1. Antimicrob. Agents Chemother. 2006, 50, 2058–2063.
- Katiyar, S.; Pfaller, M.; Edlind, T. Candida Albicans and Candida Glabrata Clinical Isolates Exhibiting Reduced Echinocandin Susceptibility. Antimicrob. Agents Chemother. 2006, 50, 2892–2894.
- Pardini, G.; De Groot, P.W.J.; Coste, A.T.; Karababa, M.; Klis, F.M.; De Koster, C.G.; Sanglard, D. The CRH Family Coding for Cell Wall Glycosylphosphatidylinositol Proteins with a Predicted Transglycosidase Domain Affects Cell Wall Organization and Virulence of Candida Albicans. J. Biol. Chem. 2006, 281, 40399–40411.
- Fonzi, W.A. PHR1 and PHR2 of Candida Albicans Encode Putative Glycosidases Required for Proper Cross-Linking of β-1,3- and β-1,6-Glucans. J. Bacteriol. 1999, 181, 7070–7079.
- Yu, Z.W.; Quinn, P.J. Solvation Effects of Dimethyl Sulphoxide on the Structure of Phospholipid Bilayers. Biophys. Chem. 1998, 70, 35–39.
- De Cesare, G.B.; Hafez, A.; Stead, D.; Llorens, C.; Munro, C.A. Biomarkers of Caspofungin Resistance in Candida Albicans Isolates: A Proteomic Approach. Virulence 2022, 13, 1005–1018.
- Caplan, T.; Polvi, E.J.; Xie, J.L.; Buckhalter, S.; Leach, M.D.; Robbins, N.; Cowen, L.E. Functional Genomic Screening Reveals Core Modulators of Echinocandin Stress Responses in Candida Albicans. Cell Rep. 2018, 23, 2292–2298.
- Kelly, J.; Kavanagh, K. Proteomic Analysis of Proteins Released from Growth-Arrested Candida Albicans Following Exposure to Caspofungin. Med. Mycol. 2010, 48, 598–605.
- Lafayette, S.L.; Collins, C.; Zaas, A.K.; Schell, W.A.; Betancourt-Quiroz, M.; Leslie Gunatilaka, A.A.; Perfect, J.R.; Cowen, L.E. PKC Signaling Regulates Drug Resistance of the Fungal Pathogen Candida Albicans via Circuitry Comprised of Mkc1, Calcineurin, and Hsp90. PLoS Pathog. 2010, 6, e1001069.
- Cowen, L.E.; Steinbach, W.J. Stress, Drugs, and Evolution: The Role of Cellular Signaling in Fungal Drug Resistance. Eukaryot. Cell 2008, 7, 747–764.
- Erbs, P.; Exinger, F.; Jund, R. Characterization of the Saccaromyces Cerevisiae FCY1 Gene Encoding Cytosine Deaminase and Its Homologue FCA1 of Candida Albicans. Curr. Genet. 1997, 31, 1–6.
- McManus, B.A.; Moran, G.P.; Higgins, J.A.; Sullivan, D.J.; Coleman, D.C. A Ser29Leu Substitution in the Cytosine Deaminase Fca1p Is Responsible for Clade-Specific Flucytosine Resistance in Candida Dubliniensis. Antimicrob. Agents Chemother. 2009, 53, 4678–4685.
- Lestrade, P.P.; Bentvelsen, R.G.; Schauwvlieghe, A.F.A.D.; Schalekamp, S.; Van Der Velden, W.J.F.M.; Kuiper, E.J.; Van Paassen, J.; Van Der Hoven, B.; Van Der Lee, H.A.; Melchers, W.J.G.; et al. Voriconazole Resistance and Mortality in Invasive Aspergillosis: A Multicenter Retrospective Cohort Study. Clin. Infect. Dis. 2019, 68, 1463–1471.
- Jund, R.; Lacroute, F. Genetic and Physiological Aspects of Resistance to 5-Fluoropyrimidines in Saccharomyces Cerevisiae. J. Bacteriol. 1970, 102, 607–615.
- Chevallier, M.R.; Jund, R.; Lacroute, F. Characterization of Cytosine Permeation in Saccharomyces Cerevisiae. J. Bacteriol. 1975, 122, 629–641.
- Kern, L.; de Montigny, J.; Lacroute, F.; Jund, R. Regulation of the Pyrimidine Salvage Pathway by the FUR1 Gene Product of Saccharomyces Cerevisiae. Curr. Genet. 1991, 19, 333–337.
- Chandra, J.; Kuhn, D.M.; Mukherjee, P.K.; Hoyer, L.L.; McCormick, T.; Ghannoum, M.A. Biofilm Formation by the Fungal Pathogen Candida Albicans: Development, Architecture, and Drug Resistance. J. Bacteriol. 2001, 183, 5385–5394.
- Rajendran, R.; Sherry, L.; Nile, C.J.; Sherriff, A.; Johnson, E.M.; Hanson, M.F.; Williams, C.; Munro, C.A.; Jones, B.J.; Ramage, G. Biofilm Formation Is a Risk Factor for Mortality in Patients with Candida Albicans Bloodstream Infection-Scotland, 2012–2013. Clin. Microbiol. Infect. 2016, 22, 87–93.
- Ramage, G.; Vande Walle, K.; Wickes, B.L.; López-Ribot, J.L. Standardized Method for in Vitro Antifungal Susceptibility Testing of Candida Albicans Biofilms. Antimicrob. Agents Chemother. 2001, 45, 2475–2479.
- Ramage, G.; Rajendran, R.; Sherry, L.; Williams, C. Fungal Biofilm Resistance. Int. J. Microbiol. 2012, 2012, 1–14.
- Fan, F.M.; Liu, Y.; Liu, Y.Q.; Lv, R.X.; Sun, W.; Ding, W.J.; Cai, Y.X.; Li, W.W.; Liu, X.; Qu, W. Candida Albicans Biofilms: Antifungal Resistance, Immune Evasion, and Emerging Therapeutic Strategies. Int. J. Antimicrob. Agents 2022, 60, 106673.
- Perumal, P.; Mekala, S.; Chaffin, W.L.J. Role for Cell Density in Antifungal Drug Resistance in Candida Albicans Biofilms. Antimicrob. Agents Chemother. 2007, 51, 2454–2463.
- Nett, J.; Lincoln, L.; Marchillo, K.; Massey, R.; Holoyda, K.; Hoff, B.; VanHandel, M.; Andes, D. Putative Role of β-1,3 Glucans in Candida Albicans Biofilm Resistance. Antimicrob. Agents Chemother. 2007, 51, 510–520.
- Nett, J.E.; Sanchez, H.; Cain, M.T.; Andes, D.R. Genetic Basis of Candida Biofilm Resistance Due to Drug-Sequestering Matrix Glucan. J. Infect. Dis. 2010, 202, 171–175.
- Tan, Y.; Ma, S.; Leonhard, M.; Moser, D.; Schneider-Stickler, B. β-1,3-Glucanase Disrupts Biofilm Formation and Increases Antifungal Susceptibility of Candida Albicans DAY185. Int. J. Biol. Macromol. 2018, 108, 942–946.
- Al-Fattani, M.A.; Douglas, L.J. Biofilm Matrix of Candida Albicans and Candida Tropicalis: Chemical Composition and Role in Drug Resistance. J. Med. Microbiol. 2006, 55, 999–1008.
- Nailis, H.; Vandenbosch, D.; Deforce, D.; Nelis, H.J.; Coenye, T. Transcriptional Response to Fluconazole and Amphotericin B in Candida Albicans Biofilms. Res. Microbiol. 2010, 161, 284–292.
- Borecká-Melkusová, S.; Moran, G.P.; Sullivan, D.J.; Kucharíková, S.; Chorvát, D.; Bujdáková, H. The Expression of Genes Involved in the Ergosterol Biosynthesis Pathway in Candida Albicans and Candida Dubliniensis Biofilms Exposed to Fluconazole. Mycoses 2009, 52, 118–128.
- Katragkou, A.; Chatzimoschou, A.; Simitsopoulou, M.; Dalakiouridou, M.; Diza-Mataftsi, E.; Tsantali, C.; Roilides, E. Differential Activities of Newer Antifungal Agents against Candida Albicans and Candida Parapsilosis Biofilms. Antimicrob. Agents Chemother. 2008, 52, 357–360.
- Rossignol, T.; Ding, C.; Guida, A.; D’Enfert, C.; Higgins, D.G.; Butler, G. Correlation between Biofilm Formation and the Hypoxic Response in Candida Parapsilosis. Eukaryot. Cell 2009, 8, 550–559.
- Knot, P.D.; Suci, P.A.; Miller, R.L.; Nelson, R.D.; Tyler, B.J. A Small Subpopulation of Blastospores in Candida Albicans Biofilms Exhibit Resistance to Amphotericin B Associated with Differential Regulation of Ergosterol and β-1,6-Glucan Pathway Genes. Antimicrob. Agents Chemother. 2006, 50, 3708–3716.
- Lepak, A.; Nett, J.; Lincoln, L.; Marchillo, K.; Andes, D. Time Course of Microbiologic Outcome and Gene Expression in Candida Albicans during and Following in Vitro and in Vivo Exposure to Fluconazole. Antimicrob. Agents Chemother. 2006, 50, 1311–1319.
- Mukherjee, P.K.; Chandra, J.; Kuhn, D.M.; Ghannoum, M.A. Mechanism of Fluconazole Resistance in Candida Albicans Biofilms: Phase-Specific Role of Efflux Pumps and Membrane Sterols. Infect. Immun. 2003, 71, 4333–4340.
- Ramage, G.; Bachmann, S.; Patterson, T.F.; Wickes, B.L.; López-Ribot, J.L. Investigation of Multidrug Efflux Pumps in Relation to Fluconazole Resistance in Candida Albicans Biofilms. J. Antimicrob. Chemother. 2002, 49, 973–980.
- Mateus, C.; Crow, S.A.; Ahearn, D.G. Adherence of Candida Albicans to Silicone Induces Immediate Enhanced Tolerance to Fluconazole. Antimicrob. Agents Chemother. 2004, 48, 3358–3366.
- Bizerra, F.C.; Nakamura, C.V.; De Poersch, C.; Estivalet Svidzinski, T.I.; Borsato Quesada, R.M.; Goldenberg, S.; Krieger, M.A.; Yamada-Ogatta, S.F. Characteristics of Biofilm Formation by Candida Tropicalis and Antifungal Resistance. FEMS Yeast Res. 2008, 8, 442–450.
- Song, J.W.; Shin, J.H.; Kee, S.J.; Kim, S.H.; Shin, M.G.; Suh, S.P.; Ryang, D.W. Expression of CgCDR1, CgCDR2, and CgERG11 in Candida Glabrata Biofilms Formed by Bloodstream Isolates. Med. Mycol. 2009, 47, 545–548.
- Andes, D.; Nett, J.; Oschel, P.; Albrecht, R.; Marchillo, K.; Pitula, A. Development and Characterization of an in Vivo Central Venous Catheter Candida Albicans Biofilm Model. Infect. Immun. 2004, 72, 6023–6031.
- Marichal, P.; Koymans, L.; Willemsens, S.; Bellens, D.; Verhasselt, P.; Luyten, W.; Borgers, M.; Ramaekers, F.C.S.; Odds, F.C.; Bossche, H. Vanden Contribution of Mutations in the Cytochrome P450 14α-Demethylase (Erg11p, Cyp51p) to Azole Resistance in Candida Albicans. Microbiol. 1999, 145, 2701–2713.
- Xu, Z.; Zhang, L.X.; Zhang, J.D.; Cao, Y.B.; Yu, Y.Y.; Wang, D.J.; Ying, K.; Chen, W.S.; Jiang, Y.Y. CDNA Microarray Analysis of Differential Gene Expression and Regulation in Clinically Drug-Resistant Isolates of Candida Albicans from Bone Marrow Transplanted Patients. Int. J. Med. Microbiol. 2006, 296, 421–434.
