Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Ebrahim Karami and Version 2 by Lindsay Dong.
Fetal development is a critical phase in prenatal care, demanding the timely identification of anomalies in ultrasound images to safeguard the well-being of both the unborn child and the mother. Medical imaging has played a pivotal role in detecting fetal abnormalities and malformations. However, despite significant advances in ultrasound technology, the accurate identification of irregularities in prenatal images continues to pose considerable challenges, often necessitating substantial time and expertise from medical professionals.
- fetal anomaly
- prenatal diagnosis
- machine learning
- deep learning
- ultrasonography imaging
1. Introduction
Fetal development is a critical phase in human growth, in which any abnormality can lead to significant health complications. The subjectivity and inaccuracies of medical sonographers and technicians in interpreting ultrasonography images often result in misdiagnoses [1][2][3][1,2,3]. Fetal anomalies can be defined as structural abnormalities in prenatal development that manifest in several critical anatomical sites, such as the fetal heart, central nervous system (CNS), lungs, and kidneys [4][5][4,5]. These anomalies can arise during various stages of pregnancy and can be caused by different genetics and environmental factors, or a combination of both, which are called multifactorial disorders (Figure 1) [6][7][6,7]. Ultrasound and genetic testing are two examples of prenatal screening and diagnostic tools that can help find these abnormalities at an earlier gestational age. Fetal abnormalities can have varying degrees of influence on a child’s health, from those that are easily treatable to those that result in the child’s death either during pregnancy or shortly after birth [8]. The occurrence of fetal anomalies differs across different populations. Structural anomalies in fetuses can be detected in approximately 3% of all pregnancies [9]. Ultrasound (US) is still the most commonly used method to safely screen for fetal anomalies during pregnancy, but it is mainly dependent on sonographer expertise and, therefore, is error-prone. In addition, US images sometimes lack high quality and discrete edges that can lead to inaccurate diagnosis [10][11][10,11]. Fetal development is crucial and complex, and abnormalities will significantly impact the children’s and sometimes the maternal health [12]. In this regard, the ever-increasing progress in the field of computer science has produced a wide variety of methods, such as machine learning (ML), deep learning (DL), and neural networks (NN), that are specific techniques within the broader field of artificial intelligence (AI) and have gained notable popularity in the medical field [13][14][15][16][17][18][13,14,15,16,17,18]. These methods include, but are not limited to, image classification, segmentation, detection of specific objects within images, and regression analysis. Consequently, numerous studies have been carried out on developing DL- and ML-based models for the accurate recognition of various types of prenatal abnormalities, including heart defects, CNS malformations, respiratory diseases, and renal anomalies in the context of chromosomal disorders or in the isolated forms.
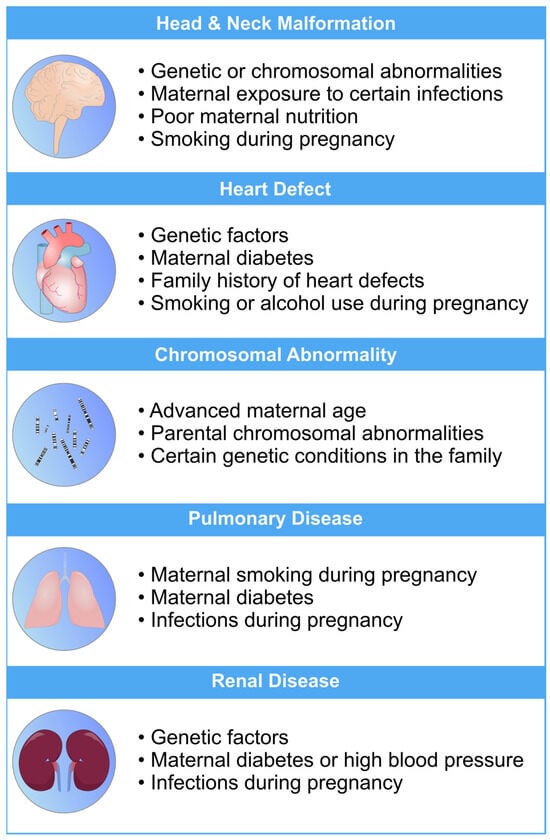
Figure 1. An overview of the most common risk factors associated with fetal abnormalities of the heart, brain, lung, and kidneys. These risk factors can have a profo bcb4und impact on the health and well-being of newborns. Limiting the exposure to these risk factors can mitigate the risk of fetal defects [19][20][21][22][23][26,27,28,29,30].
2. Methods in Machine Learning for Fetal Anomaly Detection
Machine learning (ML) is a computational technique originated from the field of computer science. In recent years, ML has been extensively used in various fields, such as medical image analysis, and has provided many valuable methods and approaches for more accurate and specific diagnoses. The field of medical image analysis is rapidly evolving, and new models and techniques are constantly emerging (Figure 23). One of the more widely used techniques in this field is deep learning (DL). A recent study has evaluated the practicality of DL-based models within clinics. They have found that AI-driven technologies can significantly help sonographers by performing disruptive tasks automatically, thus allowing technicians to focus mainly on interpreting images [24][31]. AI-based tools have great potential to lead to a paradigm shift in how we practice medicine. Many researchers have now constructed ML- and DL-based models to use in applications ranging from evaluating gestational age [25][32] to the simultaneous anomaly detection of fetal organs, which will be discussed in more detail in the following sections.
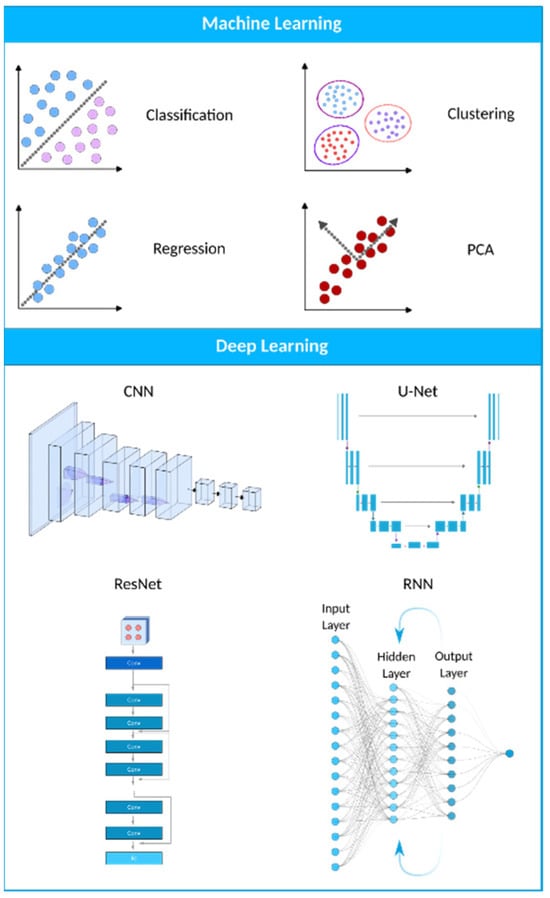

Figure 23. A visual representation of the AI landscape with three primary subsections: artificial intelligence, machine learning, and deep learning. The figure highlights four deep learning models (CNN, U-Net, ResNet, and RNN) and four machine learning algorithms (classification, clustering, PCA, and regression) as key components within these domains.
3. Applications of Machine Learning in Fetal Anomaly Detection
To fully appreciate the role of machine learning in the diagnosis of fetal abnormalities, it is necessary to first become familiar with the standard imaging technique that serves as the foundation of this diagnostic procedure. In comparison to computed tomography (CT) and magnetic resonance imaging (MRI), US imaging is the preferable method since it allows for real-time, cost-effective prenatal examination without the use of ionizing radiation. The standard procedure for fetal anomaly detection is typically a multi-step process, starting with the identification and interpretation of the sonographic images (Figure 36a). The initial scans are obtained in the first trimester, followed by a detailed anatomic survey in the second trimester. This survey involves the examination of multiple fetal organ systems and structures like the heart, brain, lungs, and kidneys, among others. Following this, the images are analyzed, pre-processed for any potential noise and errors, and finally fed into ML-based models for the detection of abnormalities or deviations from the normal developmental patterns (Figure 36b,c). ML can significantly streamline this process by automating the initial analysis and potentially identifying abnormalities with greater accuracy and speed than traditional manual interpretation.
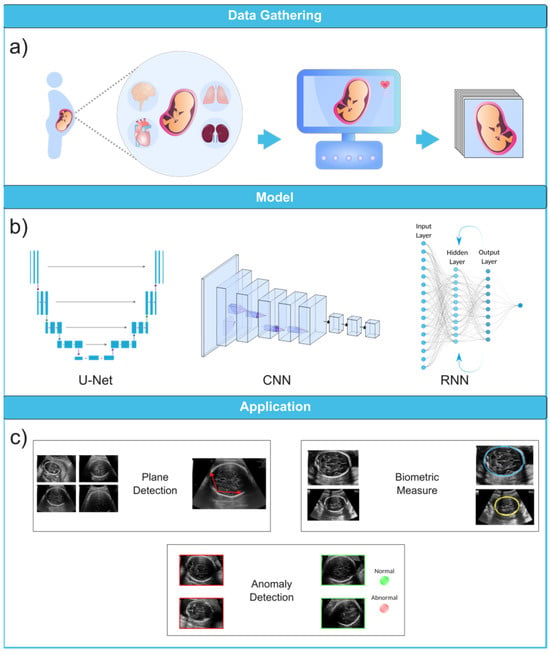

Figure 36. Ultrasound image analysis pipeline. (a) In this initial phase, ultrasound imaging is performed on a pregnant woman to identify potential fetal organ abnormalities. (b) This section presents a variety of deep learning models designed for different ultrasound image analysis task, such as CNN, U-Net, and RNN. (c) This section demonstrates the wide-ranging applications facilitated by deep learning models, including biometric measurements (e.g., head circumference), standard plane identification, and detection of fetal anomalies. Ultrasound images were obtained from the following dataset on the Kaggle (https://www.kaggle.com/datasets/rahimalargo/fetalultrasoundbrain, accessed on 1 August 2023).
3.1. Ultrasound Imaging
US imaging provides a real-time, low-cost prenatal evaluation with the additional advantages of being radiation-free and noninvasive in comparison to CT and MRI [26][59]. During a US exam, a transducer probe is placed against the mother’s abdomen and moved to visualize fetal structures. The probe transmits high-frequency sound waves, which are reflected to produce two-dimensional grayscale images representing tissue planes. The US machine calculates the time interval between transmitted and reflected waves to localize anatomical structures. Repeated pulses and reflections generate real-time visualization of the fetus. The US can capture standard views such as the four-chamber heart, profile, lips, brain, spine, and extremities [27][28][29][60,61,62]. Fetal standard planes in US imaging refer to specific anatomical views to assess fetal development. They provide a standardized orientation for evaluating different structures and measurements in the fetus, aiding in diagnosing potential abnormalities or monitoring the growth and well-being of the developing baby during pregnancy. Thus, the automatic recognition of standard planes in fetal US images is an effective method for diagnosing fetal anomalies.
Until now, numerous studies have been conducted to find the best models and approaches for reliable US image and video segmentation [30][31][32][33][66,67,68,69]. The evaluation of fetal health is the most common application of ultrasound technology. In particular, ultrasound is used to monitor the development of a fetus and detect any abnormalities early on. Placenta anomalies, growth restrictions, and structural defects all fall into this category. Due to their improved pattern recognition skills, DL models such as CNN have proven to be effective in the detection of abnormalities (Figure 47).
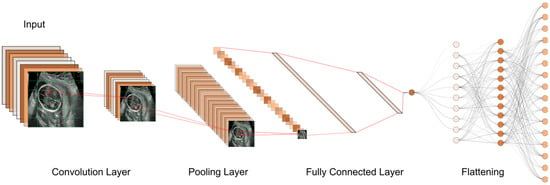

Figure 47. The typical workflow of a CNN for ultrasound image analysis. Convolution Layer: This displays the initial layer where input images are processed using convolution operations to extract features. Pooling Layer: This illustrates the subsequent layer where pooling operations (e.g., max-pooling) are applied to reduce spatial dimensions and retain important information. Fully Connected Layer: This shows the layer responsible for connecting the extracted features to make classification decisions or predictions. Flattening: This represents the process of converting the output from the previous layers into a one-dimensional vector for further processing.
3.1.1. First Trimester
First-trimester US imaging is typically performed between 11 and 13 weeks of gestation [34][77]. Its primary uses are to confirm pregnancy viability, determine gestational age, evaluate multiple gestations, and screen for significant fetal anomalies such as neural tube defects, abdominal wall defects, cardiac anomalies, nuchal translucency (NT), and some significant fetal brain abnormalities [35][36][78,79]. An abnormal NT measurement (≥3.5 mm (>p99)) during the first-trimester US can strongly predict the risk of chromosomal abnormalities and even congenital heart defects [37][38][39][80,81,82].
3.1.2. Second Trimester
Second-trimester US imaging is commonly performed between 18 and 22 weeks of gestation. The primary aim is a detailed anatomical survey to evaluate fetal growth and full screening for structural abnormalities and placental growth and status. The fetal anatomy scan assesses the brain, face, spine, heart, lungs, abdomen, kidneys, and extremities [40][83]. The second-trimester US has high detection rates for major fetal anomalies if performed by a qualified expert. The appropriateness criteria provide screening recommendations for fetuses in the second and third trimesters with varying risk levels [41][84].
3.1.3. Third Trimester
Third-trimester US imaging is often performed around 28–32 weeks of gestation to re-confirm fetal growth and position, screen for anomalies that may have developed since the prior scan, and make further assessments on the placental location and growth. It was found that fetal anomalies can be discovered in 1/300 pregnancies during routine third-trimester ultrasounds [42][85]. While US is valuable for prenatal screening, it does have limitations. The imaging quality can be impaired by the maternal body environment, fetal position, shadowing from bones, and low amniotic fluid volume [43][44][86,87]. Interpretation requires extensive training and is subject to human error. A computerized analysis of US images using ML offers the potential to overcome some human limitations. ML methods aim to improve screening accuracy and standardize interpretation by applying AI to analyze US data. These models can be trained to identify anomalies in poor-quality scans and detect subtle or complex patterns that may be missed by the technicians. However, further research is still needed to fully integrate ML into clinics and medical workflows.3.2. Diagnosis of Fetal Abnormalities
3.2.1. Congenital Heart Diseases
Congenital heart diseases (CHDs) are classified as common and severe congenital malformations in fetuses, occurring in approximately 6 to 13 out of every 1000 cases [45][88]. Although, CHDs may have no prenatal symptoms, they may result in significant morbidities, and even death, later in life. Since heart defects are the most common fetal anomalies among fetuses, research interest in this matter is consequently higher than other types of defects. Evaluating the cardiac function of a fetus is challenging due to the factors such as the fetus’s constant movement, rapid heart rate, small size, limited access, and insufficient expertise in fetal echocardiography among some sonographers, which makes the identification of complex abnormal heart structures difficult and prone to errors [46][47][48][89,90,91]. Fetal echocardiography was introduced about 25 years ago and now needs to incorporate advanced technologies. The inability to identify CHD during prenatal screening is more strongly influenced by a deficiency in adaptation skills during the performance of the SAS test than by situational variables like body mass index or fetal position. The cardiac images exhibited a considerably higher frequency of insufficient quality in undiscovered instances as compared to identified ones. In spite of the satisfactory image quality, CHD was undetected in 31% of instances. Furthermore, it is worth noting that in 20% of instances when CHD went undiscovered, the condition was not visually apparent despite the presence of high-quality images [49][92]. Echocardiography, a specialized US technique, remains the primary and essential method for early detection of fetal cardiac abnormalities and mortality risk, aimed at identifying congenital heart defects before birth. It is extensively employed during pregnancy, and the obtained images can be used to train DL models like CNN to automate and enhance the identification of abnormalities [50][93]. An echocardiogram consists of a detailed US test of the fetal heart, performed prenatally; utilizing AI for analyzing echocardiograms holds promise in advancing prenatal diagnosis and improving heart defect screening [51][94]. While GANs have demonstrated their effectiveness in anomaly detection and generative modeling, it is possible to enhance their analytical performance for intricate tasks like fetal echocardiography assessment by training an ensemble of multiple neural networks and integrating their predictions. The use of an ensemble of neural networks involves the integration of different neural networks in order to address certain machine-learning objectives. The key concept is that an ensemble of multiple neural networks would typically exhibit greater performance compared to any individual network. The four-chamber view facilitates the assessment of cardiac chamber size and the septum. In contrast, the left ventricular outflow tract view offers a visualization of the aortic valve and root. The right ventricular outflow tract view provides insight into the pulmonary valve and artery, and the three-vessel view confirms normal anatomy by showcasing the pulmonary artery, aorta, and superior vena cava. Zhou et al. [52][97] introduced a category attention network aimed at simultaneous image segmentation for the four-chamber view. They modified the SOLOv2 model for object instance segmentation. However, SOLOv2 encounters a potential misclassification issue with grids within divisions containing pixels from different instance categories. This discrepancy arises because the category score of a grid might erroneously surpass that of surrounding grids, which affects the final quality of instance segmentation. Certain image portions would become intertwined, leading to challenges in accurate object classification. To address this, the researchers integrated a “category attention module” (CAM) into SOLOv2, creating CA-ISNet. The CAM analyzes various image sections, aiding in accurately determining object categories. The proposed CA-ISNet model underwent training using a dataset of 319 images encompassing the four cardiac chambers of the fetuses.3.2.2. Head and Neck Anomalies
The development of the fetal brain is the most essential process that takes place during the 18–21 weeks of pregnancy. Any abnormalities in the fetal brain can have severe effects on various functionalities of the brain, such as cognitive function, motor skills, language development, cortical maturation, and learning capabilities [53][54][111,112]. Thus, a precise anomaly detection method is of the utmost importance. Currently, US is still the most commonly used method to initially examine the development of the fetal brain for any fetal anomalies during pregnancy. During the 18- to 21-week pregnancy period, US imaging is used to measure the cerebrum, midbrain, cerebellum, brainstem, and other regions of the brain as part of the screening for fetal abnormalities [55][56][113,114]. To detect fetal brain abnormalities, Sreelakshmy et al. developed a model (ReU-Net) based on U-Net and ResNet for the segmentation of fetuses’ cerebellum using 740 fetal brain US images [57][115]. The cerebellum is an essential part of the brain that plays a crucial role in motor control, coordination, and balance. The fetal cerebellum can be seen and distinguished from other parts of the brain in US images, which makes it relatively easy for technicians to examine it during scans and, consequently, for researchers to employ DL-based models for the segmentation of the obtained images. Moreover, ResNet is a popular model frequently used for medical image segmentation, and it offers to skip connections to address the vanishing gradient problem. More specifically, in deep networks, gradients that are used to guide the weight information update for layers can become smaller and smaller as they are multiplied at each layer, and they will eventually reach close to zero. This makes the network struggle to learn complex patterns from images, which is essential in medical image processing. Besides using ResNets, Sreelakshmy et al. also employed the Wiener filter, which reduces unwanted noises in most US images. As a result, their ReU-Net model achieved 94% and 91% for precision rate and DICE, respectively. Singh et al. also used the ResNet model in conjunction with U-Nets to automate the cerebellum segmentation procedure. However, in this study, by including residual blocks and using dilation convolution in the last two layers, they were able to improve cerebellar segmentation from noisy US images [58][116]. The subcortical volume development in a fetus is a crucial aspect to monitor during pregnancy. Hesse et al. constructed a CNN-based model for an automated segmentation of subcortical structures in 537 3D US images [59][117]. One important aspect of this research is the use of few-shot learning to train the CNN using relatively few manually annotated data (in this case, only nine). Few-shot learning is a machine learning paradigm characterized by the training of a model to perform various tasks using a very restricted amount of data. This quantity is often significantly smaller than what is typically required by conventional machine learning approaches. The basic goal of few-shot learning is to make models flexible and capable of doing tasks that would otherwise need extensive labeled data collection, which can be either time-consuming or expensive. Cystic hygroma is an abnormal growth that frequently occurs in the fetal nuchal area, within the posterior triangle of the neck. This growth originates from a lymphatic system abnormality, which develops from jugular-lymphatic blockage in 1 in every 285 fetuses [60][118]. The diagnosis of cystic hygroma is made with an evaluation of the NT thickness. Studies have also shown the connection between cystic hygroma and chromosomal abnormalities in first-trimester screenings [61][119]. In this concern, a CNN model called DenseNet was trained by Walker et al. on a dataset that included 289 sagittal fetal US images (129 images were from cystic hygroma cases, and 160 were from normal NT controls) in order to diagnose cystic hygroma in the first-trimester US images. The model was used to classify images as either “normal” or “cystic hygroma”, with an overall accuracy of 93% [62][120]. To perform US in order to look for abnormalities in the brains of prenatal fetuses, the standard planes of fetal brain are commonly used. However, fetal head plane detection is a subjective procedure, and consequently, prone to errors and mistakes by technicians. Recently, a study was conducted to automate fetal head plane detection by constructing a multi-task learning framework with regional CNNs (R-CNN). This MF R-CNN model was able to accurately locate the six fetal anatomical structures and perform a quality assessment for US images [63][124]. Based on the same dataset provided by Xie et al. [64][127], another study was conducted to develop a computer-aided framework for diagnosing fetal brain anomalies. Craniocerebral regions of fetal head images were first extracted using a DCNN with U-Nets and a VGG-Net network, and then classified into normal and abnormal categories. In small datasets, using VGG networks can lead to overfitting because of the large number of parameters available in these models. However, they used this model on a large dataset of US images and achieved an overall accuracy of 91.5%. In addition, the researchers implemented class activation mapping (CAM) to localize lesions and provide visual evidence for diagnosing abnormal cases, which can make them visually comprehensive for non-expert technicians. However, the IoU value of the predicted lesions was too low, and thus, more advanced object detection techniques are required for a more precise localization [65][128].3.2.3. Respiratory Diseases
The development and function of the lungs are crucial for the well-being and survival of fetuses. Malformations caused by underdevelopment or abnormalities inside the lung structure will lead to serious health issues and even death in newborns. For example, neonatal respiratory morbidity (NRM), such as respiratory distress syndrome or transient tachypnea of the newborn, is often seen when a fetus’ lungs are not fully developed, and it is still a major cause of morbidity and death [66][138]. Immature fetal lungs are closely linked to the respiratory complications experienced by newborns [67][139]. In addition, fetal lung lesions are estimated to manifest in around 1 in 15,000 live births, and are believed to originate from a range of abnormalities associated with fetal lung airway malformation [68][140]. In this case, the random undersampling with AdaBoost (RUSBoost) model was developed using extracted features from fetal lung images to predict NRM. However, locating regions of interest within the included images was manually performed, which is time-consuming and should be automated for use in clinics. This model was able to accurately predict NRM in fetal lung images. Small sample sizes and single-source datasets were also some of its limitations [69][141].3.2.4. Chromosomal Abnormalities
Chromosomal disorders are frequently occurring genetic conditions that contribute to congenital disabilities. These disorders arise due to abnormalities in the structure or number of chromosomes in an individual’s cells, leading to significant health challenges and impairments present from birth. There are, however, various ways to detect them early on in the pregnancy.-
NT measurement, which measures the thickness of the fluid-filled space at the back of the fetus’s neck.
-
Detailed anomaly scan, a thorough US examination that checks for any structural abnormalities in fetuses.
-
Fetal echocardiography, which focuses on evaluating the fetal heart structure and function to detect cardiac anomalies.
-
Nasal bone (NB), whose absence is a valuable biomarker of Down syndrome in the first trimester of pregnancy.
