Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Camila Xu and Version 1 by Tasneem Abaza.
CircRNAs are a recently discovered class of ncRNA molecules. They are formed during the process of RNA transcript maturation. Structurally, circRNAs are covalently closed by a connection between a downstream donor and upstream acceptor RNA splice sites linked by a phosphodiester bond.
- circular RNAs (circRNAs)
- hepatocellular carcinoma (HCC)
- immunotherapy
- cytotoxic T lymphocytes
1. What Are CircRNAs?
CircRNAs are a recently discovered class of ncRNA molecules. They are formed during the process of RNA transcript maturation. Structurally, circRNAs are covalently closed by a connection between a downstream donor and upstream acceptor RNA splice sites linked by a phosphodiester bond. CircRNAs were previously regarded as splicing junk but are now recognized as functional RNA molecules [31][1]. They have expression patterns that are particular to different tissues and cell types, and they are produced from a wide variety of genes [51][2]. It is noteworthy that circRNAs are implicated in biological processes that contribute to the development and spread of cancer [52,53][3][4].
Additionally, due to their circular shape and resistance to exoribonuclease activity, circRNAs have longer half-lives than their parental linear counterparts, making it possible to detect them even when produced at low levels [41,54,55][5][6][7]. For instance, exonic circRNAs are thought to be extremely stable in cells, with most circRNAs showing half-lives of over 48 h, as opposed to an average mRNA half-life of 10 h [54,56][6][8]. These characteristics imply that circRNAs could serve as useful biomarkers for the diagnosis and prognosis of cancer patients, as previously described in [41,50][5][9].
2. Biogenesis of CircRNAs
Recent research has shown that “back-splicing”, a type of pre-mRNA splicing, is responsible for the production of circRNAs [57][10]. CircRNAs have a distinctive closed-loop structure, created by linking a downstream 5′ splice donor site and an upstream 3′ splice acceptor site, in contrast to conventional pre-mRNAs with 5′ caps and 3′ polyadenylated tails [58,59][11][12].
CircRNAs are primarily categorized into four types [60,61][13][14] based on the origin of their genomic regions: exonic circRNAs (EcircRNAs), retained-intron or exonic-intronic circRNAs (EIcircRNAs), intronic circRNAs (ciRNAs), and tRNA intronic circRNAs (tricRNAs) [62][15]. Over 80% of the circRNAs that have been discovered are EcircRNAs, and these circRNAs are mostly found in the cytoplasm [63,64][16][17]. As EcircRNAs sponge miRNAs and/or interact with RBPs, several studies have shown that EcircRNAs play significant roles in modulating the genetic expression of several coding transcripts [65,66][18][19]. EIciRNAs and ciRNAs, which compose a minor portion of circRNAs compared to EcircRNAs, are mostly found in the nucleus and, thus, can control the expression of their parental mRNAs, as shown in Figure 1 [57][10]. The following section will cover the four associated biogenesis mechanisms of circRNAs.
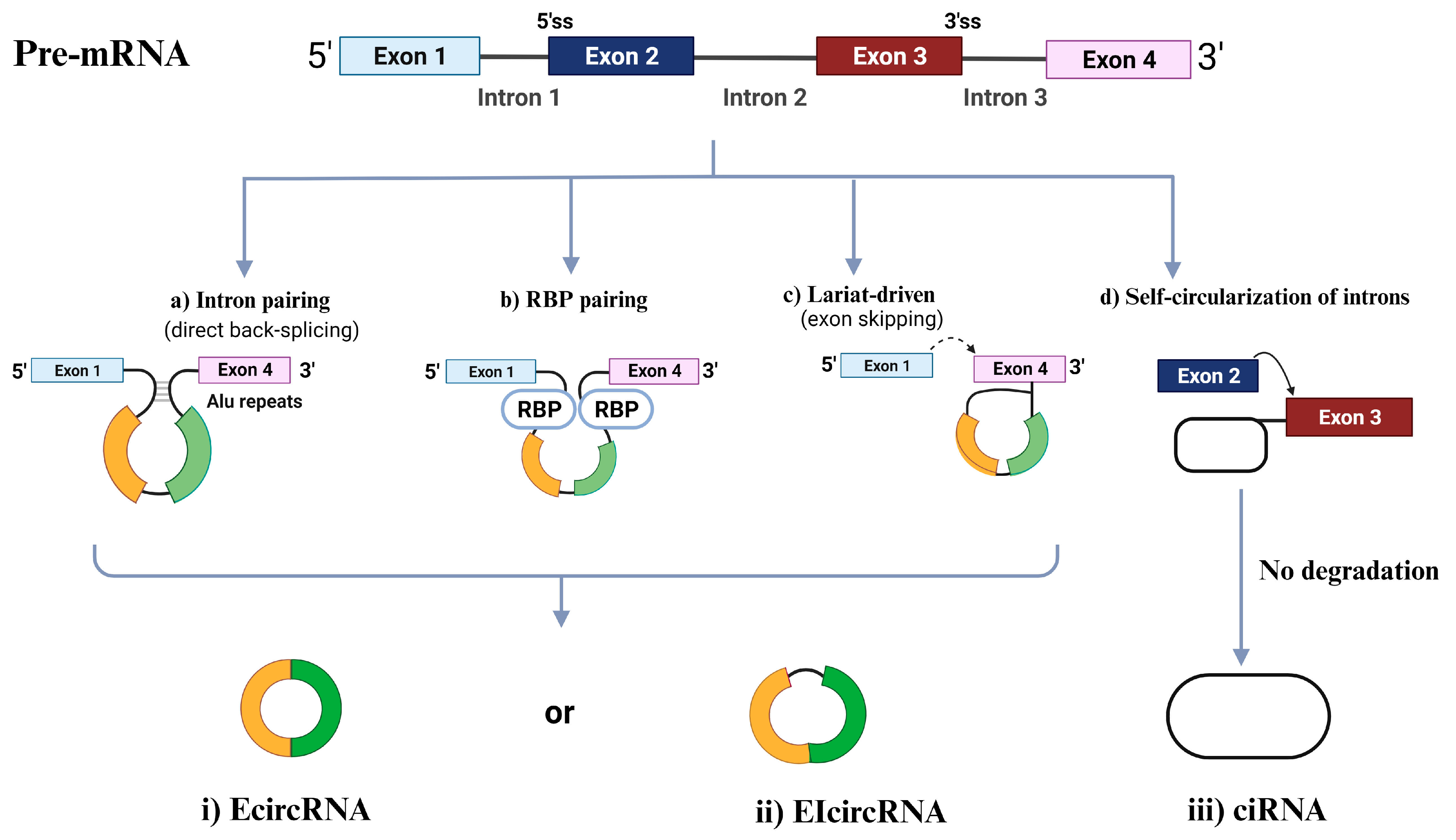
Figure 1. Circular RNA (circRNA) biogenesis. Schematic presentation of the different mechanisms of circRNAs biogenesis: intron painting (a), RBP pairing (b), Lariat driven (c), and the self-circularization of introns (d). ciRNA, intronic circRNA; EcircRNA, exonic circRNA; EIcircRNAs, exon-intron circRNA; RBP, RNA-binding protein; ss, splice site.
2.1. Intron Pairing-Driven Circularization
The most frequent circularization process of EcircRNA and EIciRNA involves “direct back-splicing”, also known as intron-pairing-driven circularization, in which a particular pre-mRNA with ALU repeats is sheared to generate an EcircRNA or an EIciRNA following reverse-base complementary pairing [56][8].
2.2. RBP-Induced Circularization
RBPs, which are thought to be trans-acting factors and include Quaking (QKI), Muscleblind (MBL), and Fused-in Sarcoma (FUS), may promote circularization by bridging similar intronic sequences [67][20]. The 3′ and 5′ termini of circularized exons can be brought into closer proximity through the dimerization of RBPs. This dimerization process also facilitates splicing by engaging with the sequences both upstream and downstream of the circularized exons [68][21].
2.3. Lariat-Induced Circularization Driven by Spliceosomes
Lariat-driven circularization, also known as the exon-skipping mechanism, occurs as pre-mRNA partially folds during transcription. This folding brings the 5′ splice site (donor site) of the upstream intron close to the 3′ splice site (receptor site) of the downstream intron, forming a circRNA through back-splicing within the folded region. The remaining exons then combine to create a linear mRNA [56][8]. Moreover, back splicing can occur post-transcriptionally or co-transcriptionally, involving either a single exon or multiple exons with intervening introns [69][22].
2.2.4. Self-Circularization of Introns
2.4. Self-Circularization of Introns
Intron self-circularization occurs when a pre-RNA contains 7 nucleotides (nt) of guanine (G) and uracil (U)-rich sequence close to one exon and an 11 nt cytosine (C)-rich sequence close to another exon. This allows the introns to avoid branching and degradation during splicing, resulting in a stable intronic lariat structure [57][10].
3. Functional Roles of CircRNAs
CircRNAs typically function as regulatory ncRNA molecules, either directly by controlling gene transcription or indirectly by modifying other regulators, such as proteins and miRNAs. Further, the term “regulatory coding RNAs” refers to a subset of circRNAs that encode short functional peptides, as shown in Figure 2 and described below [53][4].
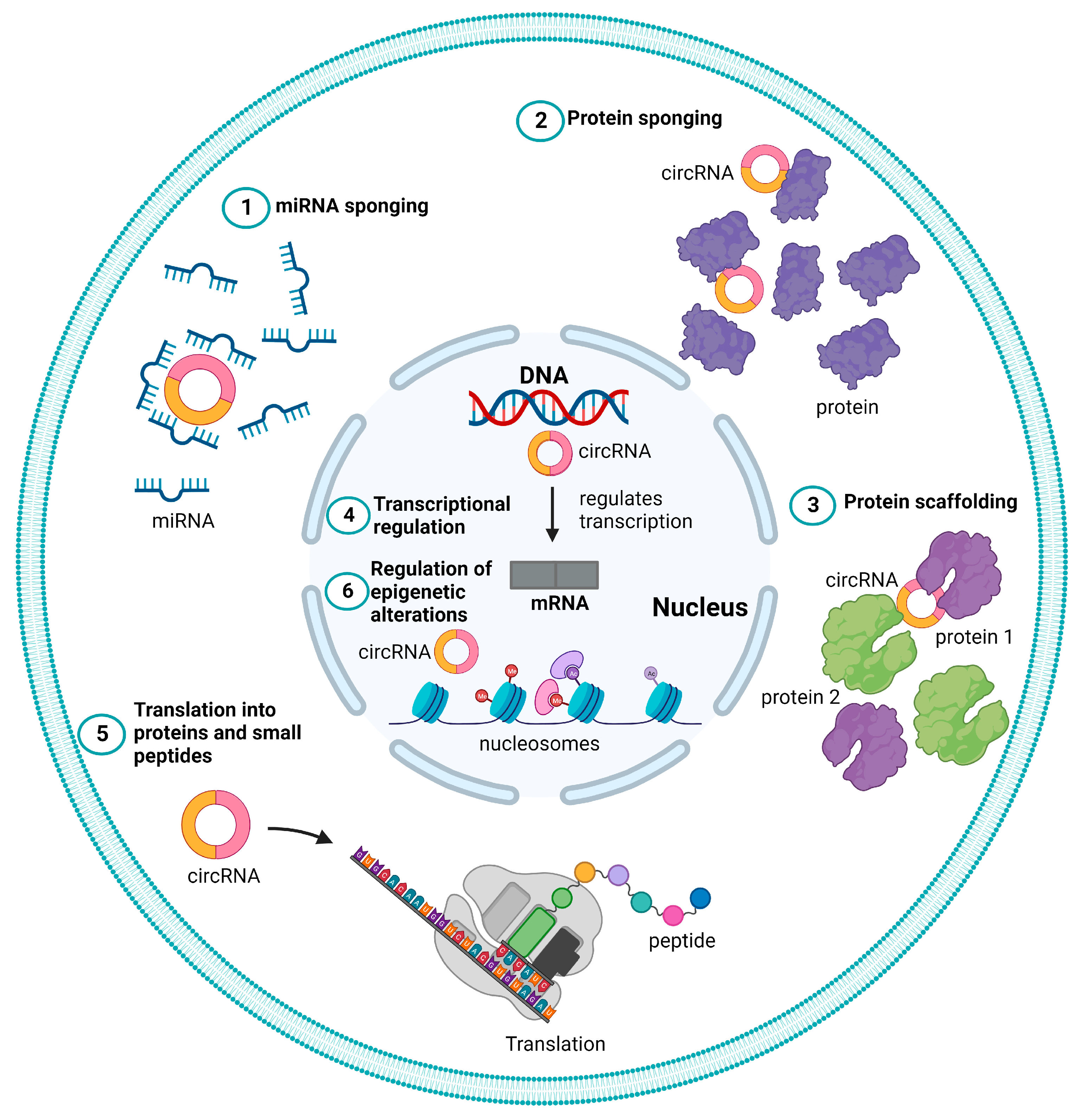
Figure 2. Different functional roles of circular RNAs (circRNAs). Schematic representation of different mechanisms of action of circRNAs represented as (1) microRNA (miRNA) sponge, (2) protein sponge or decoy (3) protein scaffolding, (4) transcriptional regulation, (5) translation to proteins, and peptide (6) regulation of epigenetic alterations.
3.1. miRNA Sponge
Some circRNAs may behave as miRNA sponges or sequesters because they include well-conserved canonical miRNA response elements (MREs) [70,71,72][23][24][25]. Some circRNAs that act as miRNA sponges can positively or adversely affect the expression of the corresponding targeted genes. Cerebellar degeneration-related protein 1 antisense (CDR1-AS or ciRS-7), a well-studied circRNA, has been linked to a variety of malignancies, including HCC and gastric cancer, as well as sponges miR-7 in embryonic zebrafish [73,74,75][26][27][28]. Indeed, a growing body of research has shown that the circRNA-miRNA-mRNA regulatory network may have significant effects on several diseases, including HCC [76,77][29][30].
For instance, circ-ZNF609 increases the expression of the myocyte-specific enhancer factor 2A (MEF2A), which improves vascular endothelial dysfunction by acting as an endogenous miR-615-5p sponge to decrease miR-615-5p activity [78][31].
According to Zhong et al., circ-MYLK can ease the inhibition of its target vascular endothelial growth factor A (VEGFA), a crucial component of the VEGFA/VEGFR2/RAS/MAPK1 signaling pathway, in addition to being associated with the stage and grade of bladder carcinoma [80][32]. By sequestering miR-143 and increasing the production of its target BCL2, increased levels of circ-UBAP2 stimulate the proliferation of osteosarcoma cells while preventing apoptosis both in vitro and in vivo [81][33]. Similarly, circ-ABCB10 has been shown by Liang et al. to sponge miR-1271, promoting proliferation and inhibiting the apoptosis of breast cancer cells [82][34].
3.2. Protein Sponge or Decoy
CircRNAs can also bind and sequester proteins using their protein-binding sites, functioning as an antagonist to impede their physiological function [83][35]. RBPs are one of the most common protein classes that can bind to circRNAs. For instance, circ-TNPO3 functions as a protein decoy for the insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) to inhibit the capacity of gastric cancer cells to proliferate [84][36]. The expression of MYC proto-oncogene, as well as bHLH transcription factor (MYC) and its target, snail family transcriptional repressor 1 (SNAI1), is inhibited when circ-TNPO3 sequesters IGF2BP3, which reduces the ability of gastric cancer cells to proliferate and metastasize [84][36]. It was also reported that circ-SIRT1 binds to the eukaryotic translation initiation factor 4A3 (EIF4A3) in colorectal cancer cell lines, preventing its inhibitory impact on epithelial–mesenchymal transition and encouraging the proliferation and invasion of colorectal cancer cell lines [86][37].
CircRNAs can also decoy proteins by attaching themselves to cellular proteins and changing how they normally carry out their physiological functions [44,87][38][39]. Circ0000079 (ciR79) inhibits the induction of fragile X-related 1 (FXR1) protein and prevents its complexation with protein kinase C iota (PRKCI), thus preventing the FXR1/PRKCI-mediated phosphorylation of glycogen synthesis kinase 3β (GSK3B) and activator protein 1 (AP-1), suppressing SNAI1 protein levels and hindering non-small cell lung cancer growth [88][40].
3.3. Protein Scaffolding
CircRNAs with enzyme and substrate binding sites are believed to serve as scaffolds that help two or more proteins to come into proximity and interact. CircFoxo3, which includes binding sites for MDM2 and p53, serves as an indicative case of this observation. In order to support the idea that circFoxo3 can serve as a protein scaffold, the mutation of these binding sites or circRNA silencing reduced the amount of p53 that an MDM2 antibody could pull down. Further research revealed that circFoxo3 promoted the ubiquitination of p53 by MDM2, which is then destroyed by the proteasome. Additionally, circACC1 forms a ternary complex with the regulatory β and γ subunits of AMP-activated protein kinase (AMPK), stabilizing and enhancing the enzymatic activity of the AMPK holoenzyme [89][41]. More circRNAs acting as scaffolds are expected to be identified in the future because of the longer half-lives of circRNAs [90][42].
3.4. Transcriptional Regulation
CircSEP3 derived from SEP3 exon 6 enhances the abundance of homologous exon 6-skipped variant by attaching to the host DNA locus and creating an RNA-DNA hybrid or R-loop, which causes transcription to pause and splicing factor recruitment [91][43]. Similarly, circSMARCA5 induces the expression of the shortened non-functional isoform by causing transcriptional termination of the SWI/SNF-related, matrix-associated, and actin-dependent regulator of chromatin, subfamily a, member 5 (SMARCA5) at exon 15 through R-loop formation [92][44]. EIciRNAs can interact with the U1 small nuclear ribonucleoprotein to increase the expression of parental genes through RNA-RNA interactions with snRNA molecules [93][45]. As lariats evade debranching, circRNAs can amass at their formation sites and enhance the activity of RNA polymerase II, resulting in the increased expression of the respective genes [57][10].
3.5. Translation to Proteins and Peptides
The ability of circRNAs to undergo translation was originally discovered by Pamudurti et al. in 2017 [94][46]. According to bioinformatics studies, some circRNAs contain an open reading frame (ORF), which indicates that they can be translated. Ribosome profiling, which can sequence ribosome-covered RNAs to track translation in vivo, has shown convincing evidence that some circRNAs comprising internal ribosome entry sites (IRES) are translated based on an IRES-dependent mechanism [94][46], whereas other circRNAs are translated independently of IRES elements. The translation of circSHPRH into the SNF2 histone linker PHD RING helicase (SHPRH)-146aa protein was demonstrated to be IRES-dependent. It was discovered that SHPRH-146aa is a tumor suppressor protein that guards against the degradation of the SHPRH full-length protein. Therefore, incorrect circSHPRH translation affects tumor malignancy [95][47].
Other circRNAs have also been discovered to encode functional peptides and proteins that have tumor-promoting or -suppressing properties [95,96,97][47][48][49]. Finally, certain circRNAs can encode peptides without the need for IRES. Protein translation is made easier, for instance, by the m6A modification. The m6A reader protein YTH N6-methyladenosine RNA binding protein F3 (YTHDF3) interacts with translation initiation factors to start protein synthesis by binding to circRNAs that include m6A modification sites [98,99,100][50][51][52].
3.6. Regulation of Epigenetic Alterations
Cancer commonly exhibits abnormal DNA methylation and histone alterations that are linked to the epigenetic regulation of gene expression [101,102][53][54]. It has been discovered that certain circRNAs control these epigenetic changes. According to Chen et al. [103][55], circFECR1 significantly reduced the amount of CpG DNA methylation in the promoter of Fli-1 proto-oncogene, ETS transcription factor (FLI1), which epigenetically activated FLI1. Through binding to the DNA methyltransferase 1 (DNMT1) promoter, circFECR1 has been shown to suppress the transcription of DNMT1, a crucial methyltransferase enzyme necessary for the upkeep of DNA methylation. Additionally, tet methylcytosine dioxygenase 1 (TET1) DNA demethylase might be attracted by circFECR1 to the FLI1 promoter and cause DNA demethylation. A component of polycomb-repressive complex 2 (PRC2), as an enhancer of zeste homolog 2 (EZH2), serves as an H3K27 methyltransferase and controls histone methylation [104,105][56][57]. Moreover, hsa-circ0020123 can upregulate EZH2 and zinc finger E-box binding homeobox 1 (ZEB1) using sponging miR-144, while circBCRC4 can enhance the expression of EZH2 by interacting with miR-101 [106,107][58][59].
4. Involvement of circRNAs in HCC Tumor Development and Progression
It has been reported that circRNAs have a fundamental role in the etiology of several human diseases, including several oncological conditions [108][60]. According to earlier investigations, circRNAs are thought to be important to the onset, development, and growth of HCC. For instance, circ0008450 induces HCC cellular proliferation, invasion, and migration and reduces apoptosis caused by sponging miR-548 [109][61]. Additionally, circRNA-104718 can similarly enhance HCC cellular proliferation, invasion, and migration and inhibit apoptosis by regulating the miRNA-218-5p/TXNDC5 axis [110][62]. The circular RNA hsa_circ_0078710 enhances cell proliferation by sequestering miR-31, resulting in the upregulation of histone deacetylase 2 (HDAC2) and cyclin-dependent kinase 2 (CDK2) expression [111][63]. Circ-ZEB1.33 facilitates the proliferation of HCC cells by modulating the miR-200a-3p/CDK6axis [112][64]. Hsa_circ_0016788 expedites HCC growth through the regulation of miR-481 and its downstream target cyclin-dependent kinase 4 (CDK4) [113][65]. Furthermore, hsa_circ_0091581 promotes the proliferation of HCC cells by elevating MYC levels, acting as a sponge for miR-526b [114][66]. Additionally, circBACH1 directly interacts with the RNA binding protein HuR, promoting the cytoplasmic accumulation of HuR, thus leading to decreased cyclin-dependent kinase inhibitor 1B (CDKN1B) expression [115][67], which influences cell cycle progression.
Pu et al. observed a significant increase in hsa_circ_0000092 expression in HCC tissues and cell lines. Depleting hsa_circ_0000092 inhibited HCC cell proliferation, migration, invasion, and angiogenesis in vitro and in vivo. This circRNA promotes HCC angiogenesis by acting as a miR-338-3p sponge, leading to increased expression of Jupiter microtubule-associated homolog 1 (JPT1), matrix metallopeptidase 9 (MMP9), and VEGFA [116][68]
Recent research highlights the pivotal roles of circRNAs in the regulation of apoptotic mechanisms within HCC. Specifically, these circRNAs target key components involved in both anti-apoptotic and pro-apoptotic signaling pathways. Notably, circ-BIRC6 exhibits significant overexpression in HCC tissue samples and correlates with the overall survival of HCC patients. Silencing circ-BIRC6 expression effectively enhances apoptosis in HCC cells by modulating BCL2 apoptosis regulator (BCL2) levels through the sequestration of miR-3918 [117][69]. Moreover, circ-0051443 displays reduced expression in HCC tissues and plasma. Exosomal circ-0051443 exerts a suppressive influence on the biological behaviors of HCC cells, primarily by promoting apoptosis through the interaction with miR-331-3p and the regulation of BCL2 antagonist/killer 1 (BAK1) [118][70]. On the other hand, certain circRNAs have inhibitory influences on the development of HCC. For instance, circADAMTS14 regulates miR-572/RCAN1, leading to the abrogation of HCC cellular hallmarks and inducing HCC cellular apoptosis machinery [119][71]; circRNA-5692 has a similar inhibitory impact on HCC progression by controlling the miR-328-5p/DAB2IP axis [120][72]. Table 1 represents a comprehensive list of all characterized oncogenic and tumor suppressor circRNAs in HCC.
Table 1.
Oncogenic and tumor suppressor circular RNAs in hepatocellular carcinoma (HCC).
| Circular RNA | Class | Molecular Targets | In Vitro/In Vivo/Ex Vivo Model | References |
|---|---|---|---|---|
| SCD-circRNA2 | Oncogenic | MAPK1, RBM3 | Huh7 HepG2 HCT-15 NCI-N87 |
[121][73] |
| circRHOT1 | Oncogenic | NR2F6 | HCC Tissues | [122][74] |
| circ-100338 | Oncogenic | MMP2, MMP9 | Hep3B HLE Huh7 BEL7402 SMCC7721 MHCC97L MHCC97H HCCLM3 HCCLM6 |
[123][75] |
| circ-0000092 | Oncogenic | miR-338-3p | Hep3B LM3 MHCC97L SK-hep1 HepG2 |
[116][68] |
| circPRMT5 | Oncogenic | miR-188-5p/HK2 axis | HCC tissues HCCLM3 SNU-387 |
[124][76] |
| circMAT2B | Oncogenic | PKM2 | HepG2 Huh7 SMMC-772 MHCC-97L MHCC-97H |
[125][77] |
| circASAP1 | Oncogenic | MAPK1 | MHCC97L MHCC97H HCCLM3 | [126][78] |
| circβ-catenin | Oncogenic | β-catenin | Huh7 | [97][49] |
| circUHRF1 | Oncogenic | UHRF1 | HepG2 HCCLM3 SMMC-7721 Huh 7 PLC/PRF/5 Hep3B |
[127][79] |
| circ-CDYL | Oncogenic | PI3K-AKT-MTORC1/β-catenin and NOTCH2 | HCCLM SMMC7721 |
[128,129][80][81] |
| circ-0046600 | Oncogenic | HIF-1α | HepG2 SK-HEP-1 |
[130][82] |
| hsa_circ_0101432 | Oncogenic | MAPK1 | Huh-7 SK-HEP-1 HepG2 HLE |
[131][83] |
| circMAN2B2 | Oncogenic | MAPK1 | HL-7702 | [132][84] |
| circPTGR1 | Oncogenic | MET | HepG2 97L LM3 |
[133][85] |
| circ-DB | Oncogenic | miR-34a, and USP7 | HepG2 Hepa 1-6 3T3L1 |
[134][86] |
| circRNA Cdr1as | Oncogenic | AFP | SMMC-7721 Bel-7402 HepG2 Hep3B Huh-7 HB611 |
[135][87] |
| circRNA PVT1 | Oncogenic | miR-203/HOXD3 pathway | SMMC-7721 Huh-7 | [136][88] |
| circPVT1 | Oncogenic | TRIM23/miR-377 axis | SNU-387 Huh-7 |
[137][89] |
| hsa_circ_0008450 | Oncogenic | EZH2 | SMMC7721 Sk-Hep-1 HepG2 Huh-7 HCCLM3 |
[138][90] |
| circ_0008450 | Oncogenic | miR-548 | HepG2 Huh-7, SMMC7721 Sk-Hep-1 HCCLM3 |
[109][61] |
| hsa_circRNA_103809 | Oncogenic | miR-377-3p/FGFR1/MAPK1 axis | MHCC97L Huh7 SK-HEP-1 Hep3B HCCLM3 |
[139][91] |
| circRNA-104718 | Oncogenic | miR-218-5p/TXNDC5 | HCC nude mice model | [110][62] |
| circMYLK | Oncogenic | miR-362-3p/Rab23 | Huh7 Hep3B |
[140][92] |
| circ-ZNF652 | Oncogenic | miR-29a-3p/GUCD1 Axis | SNU-387 Huh-7 |
[141][93] |
| circ_0000267 | Oncogenic | miR-646 | HepG2 Huh-7 SMMC7721 Sk-Hep-1 HCCLM3 |
[142][94] |
| circ-FOXP1 | Oncogenic | miR-875-3p, miR-421, SOX9 factor | SNU-387 HepG2 Hep3B Huh7 SMMC-7721 HCCLM3 |
[143][95] |
| circRNA_104075 | Oncogenic | YAP-dependent tumorigenesis through regulating HNF4a | Bel-7402 SMMC-7721 Huh7 HepG2 Hep1 Bel-7404 THLE-3 HL-7702 |
[144][96] |
| hsa_circ_101280 | Oncogenic | miR-375/JAK2 | HepG2 SNU-398 |
[145][97] |
| circRNA-101368 | Oncogenic | HMGB1/RAGE | HCCLM3 HepG2 | [146][98] |
| circ-ZEB1.33 | Oncogenic | miR-200a-3p-CDK6 | 97H Huh7 HepG2 SNU423 SNU475 L02 |
[112][64] |
| circFBLIM1 | Oncogenic | miR-346 | HCC tissues HCC mouse model |
[147][99] |
| hsa_circ_0103809 | Oncogenic | miR-490-5p/SOX2 signaling pathway | MHCC97H HepG2 Huh7 SMMC7721 SK-Hep1 |
[148][100] |
| hsa_circ_0016788 | Oncogenic | miR-486/CDK4 | HepG2 Hep3B Huh7 HCCLM3 MHCC97L |
[113][65] |
| hsa_circRBM23 | Oncogenic | miR-138 | HCC tissues HepG2 Huh7 Bel-7402 |
[149][101] |
| hsa_circ_0005075 | Oncogenic | miR-431 | SMMC-7721 | [150][102] |
| circABCC2 | Oncogenic | miR-665 | HepG2 Bel-7402 MHCC97H |
[151][103] |
| hsa_circ_100338 | Oncogenic | MTOR signaling pathway | SMMC7721 Bel-7402 Hep3B |
[152][104] |
| circ_0091581 | Oncogenic | miR-591/FOSL2 axis | THLE-2 | [153][105] |
| circPCNX | Oncogenic | miR-506 | HL-7702 SMMC-7721 HuH-7 Hep3B HepG2 |
[154][106] |
| hsa_circ_0056836 | Oncogenic | miR-766-3p/FOSL2 axis | Huh7 HepG2 SNU449 SK-HEP-1 |
[155][107] |
| circ- HOMER1 | Oncogenic | miR-1322 on CXCL6 | Sk-Hep-1 SMMC7721 HCCLM3 Huh-7 HepG2 |
[156][108] |
| circ_0091579 | Oncogenic | miR-136-5p/TRIM27 miR-1270/YAP1 miR-1225/PLCB1 |
HCCLM3 MHCC97H Huh-7 |
[157,158,159][109][110][111] |
| circ_0001955 | Oncogenic | miR-516a-5p miR-646 miR-145-5p/NRAS |
Huh-7 HepG2 SMMC-7721 Bel-7402 Hep-3B |
[160,161,162][112][113][114] |
| circTRIM33-12 | Tumor suppressor | miR-191 | HCC tissues MHCC97-L MHCC97-H LM3 |
[163][115] |
| circHIAT1 | Tumor suppressor | PTEN | Hep3B SMMC-7721 HepG2 LM3 |
[164][116] |
| circLARP4 | Tumor suppressor | miR-761/RUNX3/p53/CDKN1A pathway | Huh7 Hep3B SMMC7721 HepG2 |
[165][117] |
| circMTO1 | Tumor suppressor | miR-9-5p/NOX4 axis | HepG2 Hep3B |
[166][118] |
| circITCH | Tumor suppressor | miR-184 | Huh7 HCCLM3 SMMC-7721 MHCC97H HepG2 |
[167][119] |
| circFBXW4 | Tumor suppressor | miR-18b-3p/FBXW7 axis | LX-2 | [168][120] |
| mmu_circ_34116 | Tumor suppressor | miR-661/PTPN11 | HepG2, SNU449 | [169][121] |
| hsa_circ_0007874/cMTO1 | Tumor suppressor | miR-338-5p | HCCLM3 MHCC97-L Hep3B SMMC-7721 Huh7 Bel-7402 MHCC97-H |
[170][122] |
| circ608 | Tumor suppressor | miR-222/PINK1 | Primary hepatic stellate cells (PHSCs) from C57BL/6 mice | [171][123] |
| hsa_circ_0070963 | Tumor suppressor | miR-223-3p LEMD3 |
LX2 | [172][124] |
| hsa_circ_0004018 | Tumor suppressor | miR-626/DKK3 | Huh7 Bel7402 SNU182 Hep3B SNU449 |
[173][125] |
References
- El-Daly, S.M.; Talaat, R.M.; Braoudaki, M.; Youness, R.A.; Cho, W.C. Editorial: Recent breakthroughs in the decoding of circulating nucleic acids and their applications to human diseases. Front. Mol. Biosci. 2023, 10, 1203495.
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013, 9, e1003777.
- Dragomir, M.; Calin, G.A. Circular RNAs in Cancer—Lessons Learned From microRNAs. Front. Oncol. 2018, 8, 179.
- Li, J.; Sun, D.; Pu, W.; Wang, J.; Peng, Y. Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Trends Cancer 2020, 6, 319–336.
- Dawoud, A.; Ihab Zakaria, Z.; Hisham Rashwan, H.; Braoudaki, M.; Youness, R.A. Circular RNAs: New layer of complexity evading breast cancer heterogeneity. Noncoding RNA Res. 2023, 8, 60–74.
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461.
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e13.
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157.
- Papatsirou, M.; Artemaki, P.I.; Scorilas, A.; Kontos, C.K. The role of circular RNAs in therapy resistance of patients with solid tumors. Per Med. 2020, 17, 469–490.
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806.
- Geng, X.; Jia, Y.; Zhang, Y.; Shi, L.; Li, Q.; Zang, A.; Wang, H. Circular RNA: Biogenesis, degradation, functions and potential roles in mediating resistance to anticarcinogens. Epigenomics 2020, 12, 267–283.
- Zhang, X.O.; Dong, R.; Zhang, Y.; Zhang, J.L.; Luo, Z.; Zhang, J.; Chen, L.L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287.
- Wang, M.; Yu, F.; Li, P. Circular RNAs: Characteristics, Function and Clinical Significance in Hepatocellular Carcinoma. Cancers 2018, 10, 258.
- Bolha, L.; Ravnik-Glavac, M.; Glavac, D. Circular RNAs: Biogenesis, Function, and a Role as Possible Cancer Biomarkers. Int. J. Genom. 2017, 2017, 6218353.
- Papatsirou, M.; Artemaki, P.I.; Karousi, P.; Scorilas, A.; Kontos, C.K. Circular RNAs: Emerging Regulators of the Major Signaling Pathways Involved in Cancer Progression. Cancers 2021, 13, 2744.
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409.
- Floris, G.; Zhang, L.; Follesa, P.; Sun, T. Regulatory Role of Circular RNAs and Neurological Disorders. Mol. Neurobiol. 2017, 54, 5156–5165.
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388.
- Shen, T.; Han, M.; Wei, G.; Ni, T. An intriguing RNA species--perspectives of circularized RNA. Protein Cell 2015, 6, 871–880.
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134.
- Errichelli, L.; Dini Modigliani, S.; Laneve, P.; Colantoni, A.; Legnini, I.; Capauto, D.; Rosa, A.; De Santis, R.; Scarfo, R.; Peruzzi, G.; et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 2017, 8, 14741.
- Liang, D.; Tatomer, D.C.; Luo, Z.; Wu, H.; Yang, L.; Chen, L.L.; Cherry, S.; Wilusz, J.E. The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting. Mol. Cell 2017, 68, 940–954.e3.
- Liang, G.; Yang, Y.; Niu, G.; Tang, Z.; Li, K. Genome-wide profiling of Sus scrofa circular RNAs across nine organs and three developmental stages. DNA Res. 2017, 24, 523–535.
- Li, P.; Chen, H.; Chen, S.; Mo, X.; Li, T.; Xiao, B.; Yu, R.; Guo, J. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br. J. Cancer 2017, 116, 626–633.
- Huang, S.; Yang, B.; Chen, B.J.; Bliim, N.; Ueberham, U.; Arendt, T.; Janitz, M. The emerging role of circular RNAs in transcriptome regulation. Genomics 2017, 109, 401–407.
- Hansen, T.B.; Kjems, J.; Damgaard, C.K. Circular RNA and miR-7 in cancer. Cancer Res. 2013, 73, 5609–5612.
- Yu, L.; Gong, X.J.; Sun, L.; Zhou, Q.Y.; Lu, B.L.; Zhu, L.Y. The Circular RNA Cdr1as Act as an Oncogene in Hepatocellular Carcinoma through Targeting miR-7 Expression. PLoS ONE 2016, 11, e0158347.
- Xu, L.; Zhang, M.; Zheng, X.; Yi, P.; Lan, C.; Xu, M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2017, 143, 17–27.
- Ren, S.; Xin, Z.; Xu, Y.; Xu, J.; Wang, G. Construction and analysis of circular RNA molecular regulatory networks in liver cancer. Cell Cycle 2017, 16, 2204–2211.
- Sun, Y.; Yang, Z.; Zheng, B.; Zhang, X.H.; Zhang, M.L.; Zhao, X.S.; Zhao, H.Y.; Suzuki, T.; Wen, J.K. A Novel Regulatory Mechanism of Smooth Muscle alpha-Actin Expression by NRG-1/circACTA2/miR-548f-5p Axis. Circ. Res. 2017, 121, 628–635.
- Liu, C.; Yao, M.D.; Li, C.P.; Shan, K.; Yang, H.; Wang, J.J.; Liu, B.; Li, X.M.; Yao, J.; Jiang, Q.; et al. Silencing Of Circular RNA-ZNF609 Ameliorates Vascular Endothelial Dysfunction. Theranostics 2017, 7, 2863–2877.
- Zhong, Z.; Huang, M.; Lv, M.; He, Y.; Duan, C.; Zhang, L.; Chen, J. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017, 403, 305–317.
- Zhang, H.; Wang, G.C.; Ding, C.; Liu, P.; Wang, R.K.; Ding, W.B.; Tong, D.K.; Wu, D.J.; Li, C.; Wei, Q.; et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget 2017, 8, 61687–61697.
- Liang, H.F.; Zhang, X.Z.; Liu, B.G.; Jia, G.T.; Li, W.L. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am. J. Cancer Res. 2017, 7, 1566–1576.
- Wang, X.H.; Fang, L. Advances in circular RNAs and their roles in breast Cancer. J. Exp. Clin. Cancer Res. 2018, 37, 206.
- Yu, T.; Ran, L.; Zhao, H.; Yin, P.; Li, W.; Lin, J.; Mao, H.; Cai, D.; Ma, Q.; Pan, X.; et al. Circular RNA circ-TNPO3 suppresses metastasis of GC by acting as a protein decoy for IGF2BP3 to regulate the expression of MYC and SNAIL. Mol. Ther. Nucleic Acids 2021, 26, 649–664.
- Wang, X.; Liu, S.; Xu, B.; Liu, Y.; Kong, P.; Li, C.; Li, B. circ-SIRT1 Promotes Colorectal Cancer Proliferation and EMT by Recruiting and Binding to eIF4A3. Anal. Cell Pathol. 2021, 2021, 5739769.
- Momen-Heravi, F.; Bala, S. Emerging role of non-coding RNA in oral cancer. Cell Signal 2018, 42, 134–143.
- Huang, A.; Zheng, H.; Wu, Z.; Chen, M.; Huang, Y. Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics 2020, 10, 3503–3517.
- Chen, C.; Zhang, M.; Zhang, Y. Circ_0000079 Decoys the RNA-Binding Protein FXR1 to Interrupt Formation of the FXR1/PRCKI Complex and Decline Their Mediated Cell Invasion and Drug Resistance in NSCLC. Cell Transplant. 2020, 29, 0963689720961070.
- Li, Q.; Wang, Y.; Wu, S.; Zhou, Z.; Ding, X.; Shi, R.; Thorne, R.F.; Zhang, X.D.; Hu, W.; Wu, M. CircACC1 Regulates Assembly and Activation of AMPK Complex under Metabolic Stress. Cell Metab. 2019, 30, 157–173.e7.
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370.
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053.
- Xu, X.; Zhang, J.; Tian, Y.; Gao, Y.; Dong, X.; Chen, W.; Yuan, X.; Yin, W.; Xu, J.; Chen, K.; et al. CircRNA inhibits DNA damage repair by interacting with host gene. Mol. Cancer 2020, 19, 128.
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264.
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e7.
- Zhang, M.; Huang, N.; Yang, X.; Luo, J.; Yan, S.; Xiao, F.; Chen, W.; Gao, X.; Zhao, K.; Zhou, H.; et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 2018, 37, 1805–1814.
- Zheng, X.; Chen, L.; Zhou, Y.; Wang, Q.; Zheng, Z.; Xu, B.; Wu, C.; Zhou, Q.; Hu, W.; Wu, C.; et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol. Cancer 2019, 18, 47.
- Liang, W.C.; Wong, C.W.; Liang, P.P.; Shi, M.; Cao, Y.; Rao, S.T.; Tsui, S.K.; Waye, M.M.; Zhang, Q.; Fu, W.M.; et al. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019, 20, 84.
- Di Timoteo, G.; Dattilo, D.; Centron-Broco, A.; Colantoni, A.; Guarnacci, M.; Rossi, F.; Incarnato, D.; Oliviero, S.; Fatica, A.; Morlando, M.; et al. Modulation of circRNA Metabolism by m(6)A Modification. Cell Rep. 2020, 31, 107641.
- Lee, Y.; Choe, J.; Park, O.H.; Kim, Y.K. Molecular Mechanisms Driving mRNA Degradation by m(6)A Modification. Trends Genet. 2020, 36, 177–188.
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017, 27, 626–641.
- Bird, A. DNA methylation patterns and epigenetic memory. Genes. Dev. 2002, 16, 6–21.
- Lv, J.F.; Hu, L.; Zhuo, W.; Zhang, C.M.; Zhou, H.H.; Fan, L. Epigenetic alternations and cancer chemotherapy response. Cancer Chemother. Pharmacol. 2016, 77, 673–684.
- Chen, N.; Zhao, G.; Yan, X.; Lv, Z.; Yin, H.; Zhang, S.; Song, W.; Li, X.; Li, L.; Du, Z.; et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018, 19, 218.
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349.
- Su, M.; Xiao, Y.; Tang, J.; Wu, J.; Ma, J.; Tian, B.; Zhou, Y.; Wang, H.; Yang, D.; Liao, Q.J.; et al. Role of lncRNA and EZH2 Interaction/Regulatory Network in Lung Cancer. J. Cancer 2018, 9, 4156–4165.
- Li, B.; Xie, F.; Zheng, F.X.; Jiang, G.S.; Zeng, F.Q.; Xiao, X.Y. Overexpression of CircRNA BCRC4 regulates cell apoptosis and MicroRNA-101/EZH2 signaling in bladder cancer. Curr. Med. Sci. 2017, 37, 886–890.
- Qu, D.; Yan, B.; Xin, R.; Ma, T. A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am. J. Cancer Res. 2018, 8, 1387–1402.
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691.
- Zhang, J.; Chang, Y.; Xu, L.; Qin, L. Elevated expression of circular RNA circ_0008450 predicts dismal prognosis in hepatocellular carcinoma and regulates cell proliferation, apoptosis, and invasion via sponging miR-548p. J. Cell Biochem. 2019, 120, 9487–9494.
- Yu, J.; Yang, M.; Zhou, B.; Luo, J.; Zhang, Z.; Zhang, W.; Yan, Z. CircRNA-104718 acts as competing endogenous RNA and promotes hepatocellular carcinoma progression through microRNA-218-5p/TXNDC5 signaling pathway. Clin. Sci. 2019, 133, 1487–1503.
- Xie, B.; Zhao, Z.; Liu, Q.; Wang, X.; Ma, Z.; Li, H. CircRNA has_circ_0078710 acts as the sponge of microRNA-31 involved in hepatocellular carcinoma progression. Gene 2019, 683, 253–261.
- Gong, Y.; Mao, J.; Wu, D.; Wang, X.; Li, L.; Zhu, L.; Song, R. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int. 2018, 18, 116.
- Guan, Z.; Tan, J.; Gao, W.; Li, X.; Yang, Y.; Li, X.; Li, Y.; Wang, Q. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. J. Cell Physiol. 2018, 234, 500–508.
- Wei, X.; Zheng, W.; Tian, P.; He, Y.; Liu, H.; Peng, M.; Li, X.; Liu, X. Oncogenic hsa_circ_0091581 promotes the malignancy of HCC cell through blocking miR-526b from degrading c-MYC mRNA. Cell Cycle 2020, 19, 817–824.
- Liu, B.; Yang, G.; Wang, X.; Liu, J.; Lu, Z.; Wang, Q.; Xu, B.; Liu, Z.; Li, J. CircBACH1 (hsa_circ_0061395) promotes hepatocellular carcinoma growth by regulating p27 repression via HuR. J. Cell Physiol. 2020, 235, 6929–6941.
- Pu, J.; Wang, J.; Li, W.; Lu, Y.; Wu, X.; Long, X.; Luo, C.; Wei, H. hsa_circ_0000092 promotes hepatocellular carcinoma progression through up-regulating HN1 expression by binding to microRNA-338-3p. J. Cell Mol. Med. 2020.
- Yang, G.; Wang, X.; Liu, B.; Lu, Z.; Xu, Z.; Xiu, P.; Liu, Z.; Li, J. circ-BIRC6, a circular RNA, promotes hepatocellular carcinoma progression by targeting the miR-3918/Bcl2 axis. Cell Cycle 2019, 18, 976–989.
- Chen, W.; Quan, Y.; Fan, S.; Wang, H.; Liang, J.; Huang, L.; Chen, L.; Liu, Q.; He, P.; Ye, Y. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020, 475, 119–128.
- Song, C.; Li, D.; Liu, H.; Sun, H.; Liu, Z.; Zhang, L.; Hu, Y. The competing endogenous circular RNA ADAMTS14 suppressed hepatocellular carcinoma progression through regulating microRNA-572/regulator of calcineurin 1. J. Cell Physiol. 2019, 234, 2460–2470.
- Liu, Z.; Yu, Y.; Huang, Z.; Kong, Y.; Hu, X.; Xiao, W.; Quan, J.; Fan, X. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis. 2019, 10, 900.
- Dong, W.; Dai, Z.H.; Liu, F.C.; Guo, X.G.; Ge, C.M.; Ding, J.; Liu, H.; Yang, F. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine 2019, 45, 155–167.
- Wang, L.; Long, H.; Zheng, Q.; Bo, X.; Xiao, X.; Li, B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol. Cancer 2019, 18, 119.
- Huang, X.Y.; Huang, Z.L.; Huang, J.; Xu, B.; Huang, X.Y.; Xu, Y.H.; Zhou, J.; Tang, Z.Y. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J. Exp. Clin. Cancer Res. 2020, 39, 20.
- Ding, Z.; Guo, L.; Deng, Z.; Li, P. Circ-PRMT5 enhances the proliferation, migration and glycolysis of hepatoma cells by targeting miR-188-5p/HK2 axis. Ann. Hepatol. 2020, 19, 269–279.
- Li, Q.; Pan, X.; Zhu, D.; Deng, Z.; Jiang, R.; Wang, X. Circular RNA MAT2B Promotes Glycolysis and Malignancy of Hepatocellular Carcinoma Through the miR-338-3p/PKM2 Axis Under Hypoxic Stress. Hepatology 2019, 70, 1298–1316.
- Hu, Z.Q.; Zhou, S.L.; Li, J.; Zhou, Z.J.; Wang, P.C.; Xin, H.Y.; Mao, L.; Luo, C.B.; Yu, S.Y.; Huang, X.W.; et al. Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology 2020, 72, 906–922.
- Zhang, P.F.; Gao, C.; Huang, X.Y.; Lu, J.C.; Guo, X.J.; Shi, G.M.; Cai, J.B.; Ke, A.W. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol. Cancer 2020, 19, 110.
- Xiong, D.; He, R.; Dang, Y.; Wu, H.; Feng, Z.; Chen, G. The Latest Overview of circRNA in the Progression, Diagnosis, Prognosis, Treatment, and Drug Resistance of Hepatocellular Carcinoma. Front. Oncol. 2020, 10, 608257.
- Wei, Y.; Chen, X.; Liang, C.; Ling, Y.; Yang, X.; Ye, X.; Zhang, H.; Yang, P.; Cui, X.; Ren, Y.; et al. A Noncoding Regulatory RNAs Network Driven by Circ-CDYL Acts Specifically in the Early Stages Hepatocellular Carcinoma. Hepatology 2020, 71, 130–147.
- Zhai, Z.; Fu, Q.; Liu, C.; Zhang, X.; Jia, P.; Xia, P.; Liu, P.; Liao, S.; Qin, T.; Zhang, H. Emerging Roles Of hsa-circ-0046600 Targeting The miR-640/HIF-1alpha Signalling Pathway In The Progression Of HCC. Onco Targets Ther. 2019, 12, 9291–9302.
- Zou, H.; Xu, X.; Luo, L.; Zhang, Y.; Luo, L.; Yao, Y.; Xiang, G.; Huang, X.; Wang, G. Hsa_circ_0101432 promotes the development of hepatocellular carcinoma (HCC) by adsorbing miR-1258 and miR-622. Cell Cycle 2019, 18, 2398–2413.
- Fu, X.; Zhang, J.; He, X.; Yan, X.; Wei, J.; Huang, M.; Liu, Y.; Lin, J.; Hu, H.; Liu, L. Circular RNA MAN2B2 promotes cell proliferation of hepatocellular carcinoma cells via the miRNA-217/MAPK1 axis. J. Cancer 2020, 11, 3318–3326.
- Wang, G.; Liu, W.; Zou, Y.; Wang, G.; Deng, Y.; Luo, J.; Zhang, Y.; Li, H.; Zhang, Q.; Yang, Y.; et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine 2019, 40, 432–445.
- Zhang, H.; Deng, T.; Ge, S.; Liu, Y.; Bai, M.; Zhu, K.; Fan, Q.; Li, J.; Ning, T.; Tian, F.; et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene 2019, 38, 2844–2859.
- Su, Y.; Lv, X.; Yin, W.; Zhou, L.; Hu, Y.; Zhou, A.; Qi, F. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging 2019, 11, 8183–8203.
- Zhu, Y.; Liu, Y.; Xiao, B.; Cai, H.; Liu, M.; Ma, L.; Yin, H.; Wang, F. The circular RNA PVT1/miR-203/HOXD3 pathway promotes the progression of human hepatocellular carcinoma. Biol. Open 2019, 8, bio043687.
- Bu, N.; Dong, Z.; Zhang, L.; Zhu, W.; Wei, F.; Zheng, S. CircPVT1 Regulates Cell Proliferation, Apoptosis and Glycolysis in Hepatocellular Carcinoma via miR-377/TRIM23 Axis. Cancer Manag. Res. 2020, 12, 12945–12956.
- Lin, T.; Dai, Y.; Guo, X.; Chen, W.; Zhao, J.; Cao, L.; Wu, Z. Silencing Of hsa_circ_0008450 Represses Hepatocellular Carcinoma Progression Through Regulation Of microRNA-214-3p/EZH2 Axis. Cancer Manag. Res. 2019, 11, 9133–9143.
- Zhan, W.; Liao, X.; Chen, Z.; Li, L.; Tian, T.; Yu, L.; Wang, W.; Hu, Q. Circular RNA hsa_circRNA_103809 promoted hepatocellular carcinoma development by regulating miR-377-3p/FGFR1/ERK axis. J. Cell Physiol. 2020, 235, 1733–1745.
- Li, Z.; Hu, Y.; Zeng, Q.; Wang, H.; Yan, J.; Li, H.; Yu, Z. Circular RNA MYLK promotes hepatocellular carcinoma progression by increasing Rab23 expression by sponging miR-362-3p. Cancer Cell Int. 2019, 19, 211.
- Li, Y.; Zang, H.; Zhang, X.; Huang, G. Exosomal Circ-ZNF652 Promotes Cell Proliferation, Migration, Invasion and Glycolysis in Hepatocellular Carcinoma via miR-29a-3p/GUCD1 Axis. Cancer Manag. Res. 2020, 12, 7739–7751.
- Pan, H.; Tang, L.; Jiang, H.; Li, X.; Wang, R.; Gao, J.; Li, Q. Enhanced expression of circ_0000267 in hepatocellular carcinoma indicates poor prognosis and facilitates cell progression by sponging miR-646. J. Cell Biochem. 2019, 120, 11350–11357.
- Wang, W.; Li, Y.; Li, X.; Liu, B.; Han, S.; Li, X.; Zhang, B.; Li, J.; Sun, S. Circular RNA circ-FOXP1 induced by SOX9 promotes hepatocellular carcinoma progression via sponging miR-875-3p and miR-421. Biomed. Pharmacother. 2020, 121, 109517.
- Zhang, X.; Xu, Y.; Qian, Z.; Zheng, W.; Wu, Q.; Chen, Y.; Zhu, G.; Liu, Y.; Bian, Z.; Xu, W.; et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 2018, 9, 1091.
- Cao, S.; Wang, G.; Wang, J.; Li, C.; Zhang, L. Hsa_circ_101280 promotes hepatocellular carcinoma by regulating miR-375/JAK2. Immunol. Cell Biol. 2019, 97, 218–228.
- Li, S.; Gu, H.; Huang, Y.; Peng, Q.; Zhou, R.; Yi, P.; Chen, R.; Huang, Z.; Hu, X.; Huang, Y.; et al. Circular RNA 101368/miR-200a axis modulates the migration of hepatocellular carcinoma through HMGB1/RAGE signaling. Cell Cycle 2018, 17, 2349–2359.
- Bai, N.; Peng, E.; Qiu, X.; Lyu, N.; Zhang, Z.; Tao, Y.; Li, X.; Wang, Z. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J. Exp. Clin. Cancer Res. 2018, 37, 172.
- Cai, H.; Hu, B.; Ji, L.; Ruan, X.; Zheng, Z. Hsa_circ_0103809 promotes cell proliferation and inhibits apoptosis in hepatocellular carcinoma by targeting miR-490-5p/SOX2 signaling pathway. Am. J. Transl. Res. 2018, 10, 1690–1702.
- Wang, B.; Chen, H.; Zhang, C.; Yang, T.; Zhao, Q.; Yan, Y.; Zhang, Y.; Xu, F. Effects of hsa_circRBM23 on Hepatocellular Carcinoma Cell Viability and Migration as Produced by Regulating miR-138 Expression. Cancer Biother. Radiopharm. 2018, 33, 194–202.
- Li, M.F.; Li, Y.H.; He, Y.H.; Wang, Q.; Zhang, Y.; Li, X.F.; Meng, X.M.; Huang, C.; Li, J. Emerging roles of hsa_circ_0005075 targeting miR-431 in the progress of HCC. Biomed. Pharmacother. 2018, 99, 848–858.
- Bai, N.; Peng, E.; Xia, F.; Wang, D.; Li, X.; Li, X. CircABCC2 Regulates Hepatocellular Cancer Progression by Decoying MiR-665. J. Cancer 2019, 10, 3893–3898.
- Huang, X.Y.; Huang, Z.L.; Zhang, P.B.; Huang, X.Y.; Huang, J.; Wang, H.C.; Xu, B.; Zhou, J.; Tang, Z.Y. CircRNA-100338 Is Associated With mTOR Signaling Pathway and Poor Prognosis in Hepatocellular Carcinoma. Front. Oncol. 2019, 9, 392.
- Ji, C.; Hong, X.; Lan, B.; Lin, Y.; He, Y.; Chen, J.; Liu, X.; Ye, W.; Mo, Z.; She, Z.; et al. Circ_0091581 Promotes the Progression of Hepatocellular Carcinoma Through Targeting miR-591/FOSL2 Axis. Dig. Dis. Sci. 2021, 66, 3074–3085.
- Sun, P.; Fan, X.; Hu, X.; Fu, X.; Wei, Q.; Zang, Y. circPCNX and Pecanex Promote Hepatocellular Carcinoma Cell Viability by Inhibiting miR-506. Cancer Manag. Res. 2019, 11, 10957–10967.
- Li, Z.; Liu, Y.; Yan, J.; Zeng, Q.; Hu, Y.; Wang, H.; Li, H.; Li, J.; Yu, Z. Circular RNA hsa_circ_0056836 functions an oncogenic gene in hepatocellular carcinoma through modulating miR-766-3p/FOSL2 axis. Aging 2020, 12, 2485–2497.
- Zhao, M.; Dong, G.; Meng, Q.; Lin, S.; Li, X. Circ-HOMER1 enhances the inhibition of miR-1322 on CXCL6 to regulate the growth and aggressiveness of hepatocellular carcinoma cells. J. Cell Biochem. 2020, 121, 4440–4449.
- Liu, M.; Guo, B.; Zhang, G.; Qi, H. Circ_0091579 Knockdown Inhibited HCC Proliferation and Glutamine Metabolism Through miR-1270/YAP1 Axis. Biochem. Genet. 2023.
- Mao, Y.; Ding, Z.; Jiang, M.; Yuan, B.; Zhang, Y.; Zhang, X. Circ_0091579 exerts an oncogenic role in hepatocellular carcinoma via mediating miR-136-5p/TRIM27. Biomed. J. 2022, 45, 883–895.
- Zhang, D.; Zhang, Y.; Zhang, X.; Zhai, H.; Sun, X.; Li, Y. Circ_0091579 Serves as a Tumor-Promoting Factor in Hepatocellular Carcinoma Through miR-1225-5p/PLCB1 Axis. Dig. Dis. Sci. 2022, 67, 585–597.
- Ding, B.; Fan, W.; Lou, W. hsa_circ_0001955 Enhances In Vitro Proliferation, Migration, and Invasion of HCC Cells through miR-145-5p/NRAS Axis. Mol. Ther. Nucleic Acids 2020, 22, 445–455.
- Yao, Z.; Xu, R.; Yuan, L.; Xu, M.; Zhuang, H.; Li, Y.; Zhang, Y.; Lin, N. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis. 2019, 10, 945.
- Li, X.; Lv, J.; Hou, L.; Guo, X. Circ_0001955 Acts as a miR-646 Sponge to Promote the Proliferation, Metastasis and Angiogenesis of Hepatocellular Carcinoma. Dig. Dis. Sci. 2022, 67, 2257–2268.
- Zhang, P.F.; Wei, C.Y.; Huang, X.Y.; Peng, R.; Yang, X.; Lu, J.C.; Zhang, C.; Gao, C.; Cai, J.B.; Gao, P.T.; et al. Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol. Cancer 2019, 18, 105.
- Wang, Z.; Zhao, Y.; Wang, Y.; Jin, C. Circular RNA circHIAT1 inhibits cell growth in hepatocellular carcinoma by regulating miR-3171/PTEN axis. Biomed. Pharmacother. 2019, 116, 108932.
- Chen, Z.; Zuo, X.; Pu, L.; Zhang, Y.; Han, G.; Zhang, L.; Wu, J.; Wang, X. circLARP4 induces cellular senescence through regulating miR-761/RUNX3/p53/p21 signaling in hepatocellular carcinoma. Cancer Sci. 2019, 110, 568–581.
- Wang, J.; Tan, Q.; Wang, W.; Yu, J. Mechanism of the Regulatory Effect of Overexpression of circMTO1 on Proliferation and Apoptosis of Hepatoma Cells via miR-9-5p/NOX4 Axis. Cancer Manag. Res. 2020, 12, 3915–3925.
- Guo, X.; Wang, Z.; Deng, X.; Lu, Y.; Huang, X.; Lin, J.; Lan, X.; Su, Q.; Wang, C. Circular RNA CircITCH (has-circ-0001141) suppresses hepatocellular carcinoma (HCC) progression by sponging miR-184. Cell Cycle 2022, 21, 1557–1577.
- Chen, X.; Li, H.D.; Bu, F.T.; Li, X.F.; Chen, Y.; Zhu, S.; Wang, J.N.; Chen, S.Y.; Sun, Y.Y.; Pan, X.Y.; et al. Circular RNA circFBXW4 suppresses hepatic fibrosis via targeting the miR-18b-3p/FBXW7 axis. Theranostics 2020, 10, 4851–4870.
- Matboli, M.; Hassan, M.K.; Ali, M.A.; Mansour, M.T.; Elsayed, W.; Atteya, R.; Aly, H.S.; Meteini, M.E.; Elghazaly, H.; El-Khamisy, S.; et al. Impact of circ-0000221 in the Pathogenesis of Hepatocellular via Modulation of miR-661-PTPN11 mRNA Axis. Pharmaceutics 2022, 14, 138.
- Shen, H.; Li, H.; Zhou, J. Circular RNA hsa_circ_0032683 inhibits the progression of hepatocellular carcinoma by sponging microRNA-338-5p. Bioengineered 2022, 13, 2321–2335.
- Xu, Z.X.; Li, J.Z.; Li, Q.; Xu, M.Y.; Li, H.Y. CircRNA608-microRNA222-PINK1 axis regulates the mitophagy of hepatic stellate cells in NASH related fibrosis. Biochem. Biophys. Res. Commun. 2022, 610, 35–42.
- Ji, D.; Chen, G.F.; Wang, J.C.; Ji, S.H.; Wu, X.W.; Lu, X.J.; Chen, J.L.; Li, J.T. Hsa_circ_0070963 inhibits liver fibrosis via regulation of miR-223-3p and LEMD3. Aging 2020, 12, 1643–1655.
- Zhu, P.; Liang, H.; Huang, X.; Zeng, Q.; Liu, Y.; Lv, J.; Ming, L. Circular RNA Hsa_circ_0004018 Inhibits Wnt/beta-Catenin Signaling Pathway by Targeting microRNA-626/DKK3 in Hepatocellular Carcinoma. Onco Targets Ther. 2020, 13, 9351–9364.
More
