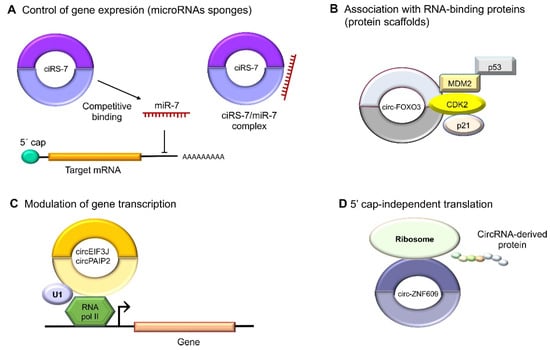
Figure 3. Molecular functions of circular RNAs. (
A) CircRNAs act indirectly on gene expression as microRNA sponges: for example, oncogenic ciRS-7 (CDR1as) competitively binds to tumor suppressor miR-7, since it contains miRNA response elements (MREs). (
B) CircRNAs can also bind to RBPs such as circ-FOXO3, which interacts with MDM2 (murine double-minute 2) to degrade p53 by ubiquitination, forming a ternary complex with CDK2 and p21. (
C) The nuclear circRNAs that conserve the intronic sequences of their parental genes, circEIF3J and circPAIP2, to improve those genes’ cis expressions could associate with RNA polymerase II, which would improve their functions in a U1 snRNP-dependent manner. (
D) Multiple circRNAs contain internal ribosome entry sites (IRESs), which suggests that they can be translated into peptides or proteins in a 5′ cap-independent manner, as in the case of circ-ZNF609, the translation of which plays an important role in myogenesis.
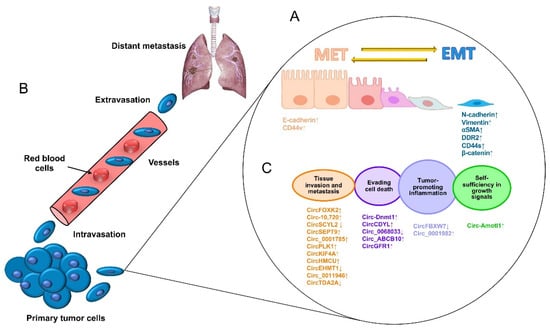
4. Circular RNAs Functions in Triple-Negative Breast Cancer
CircRNAs Sponge Relevant MicroRNAs Involved in Tumor Progression and Metastasis
At least 60% of the human transcriptome belongs to ncRNAs that participate in biological processes as differential regulators with pathological impacts
[20][38]. The role of circRNAs in breast cancer, especially in the triple-negative subtype, has recently been demonstrated. Some studies have shown that circRNAs act mainly through the kidnapping of microRNAs, which regulates important oncogenic and tumor-suppressor genes involved in the regulation of cancer hallmarks. Moreover, breast cancer metastasis has been associated with alterations in the expressions of multiple circRNAs and microRNAs. On the other hand, circBCBM1 was detected in the brain as a promoter of metastasis through the modulation of the miR-125a/BRD4 axis
[21][40]. Another relevant circular RNA is circZEB1, which has been markedly overexpressed in TNBC tissues and cell lines, promoting proliferation, and reducing apoptosis through the miR-448/eEF2K axis
[22][41].
Similarly, the overexpression of circBACH2 has been reported to facilitate the EMT, activating cancer cell invasion and migration as well as proliferation through the binding of both tumor suppressors miR-186-5p and miR-548c-3p, thus promoting the expression of the oncogenic chemokine receptor CXC type 4, or CXCR4. Moreover, circBACH2 was found expressed on the surfaces of various types of cancer cells; thus, its depletion has resulted in the suppression of the malignant progression of TNBC cells
[23][50].
Intriguingly, Li et al. discovered that a circRNA derived from the HER2 gene, circ-HER2, was expressed in TNBC even when its classification indicated the absence of the HER2 receptor. The expression of the circ-HER2 contributed to homo/hetero EGFR/HER3 dimerization, which originated the sustained phosphorylation of AKT that promotes proliferation, invasion, and metastasis. The circ-HER2 expression correlated with poor prognosis in patients
[24][53].
Remarkably, circRNAs have been described to not only promote the metastasis of cancer cells but possibly also play the opposite role: strong tumor suppression. For example, the overexpression of circCDYL has promoted apoptosis and inhibited cell proliferation through regulation of the miR-190a-3p/TP53INP1 axis, therefore upregulating the tumor-suppressing TP53INP1 protein in TNBC
[25][59].
Finally, circRNA molecules could be derived from diverse genomic regions, as in the case of hsa_circ_0091074, which is generated from a genomic region of the X-specific inactive transcript (XIST) and binds to miR-1297, partially blocking the antitumor action of its target microRNA and resulting in the overexpression of the TAZ protein and positive regulation of the cell cycle in TNBC cells
[26][62].
5. Circular RNAs Exported in Exosomes in TNBC
In recent decades, exosomes have become an important topic of study given their important roles in the regulation of cancer hallmarks. Exosomes are nanometric vesicles (40–100 nm in diameter) of endocytic origin that share a similar topology and lipid composition with the plasma membrane. From inside, a wide variety of cargo molecules, such as nucleic acids, proteins, and enzymes capable of modulating cellular activities in recipient cells through the transfer of functional genetic information, can be found
[27][68]. Exosome biogenesis is mediated by two pathways that classify the proteins that they contain. The endosomal sorting complex transport (ESCRT)-dependent pathway is the best-characterized mechanism, involving four large protein complexes and more than 30 proteins. The ESCRT-independent pathway involves the inhibition of a neutral sphingomyelinase (nSMase) that is necessary to hydrolyze sphingomyelin and originate ceramide
[28][69]. Exosomes are formed by budding from the membranes of multivesicular bodies (MVBs) from late endosomes: invaginations, called intraluminal vesicles (ILVs), that contain cytosolic components. MVBs can fuse with lysosomes for their degradation or may follow the endocytic pathway for the generation of exosomes
[29][70].
Exosomal circRNAs are known to play an important role in cancer biology, as they can be taken up by neighboring or distant cells and affect many aspects of the physiological and pathological conditions of recipient cells. Recently, exosomes were discovered to be enriched with circRNA molecules, as demonstrated by Li et al. More than 1000 circRNAs have been identified in human serum exosomes. Notably, the circRNA content in exosomes is regulated by changes in the levels of the associated microRNAs in the producer cells
[30][71]. On the other hand, Yang et al. found that circPSMA1 was overexpressed in serum exosomes from TNBC patients.
6. Circular RNAs Regulation of Resistance to Chemo/Radiotherapy
The expression of circRNAs related to the doxorubicin (DOX) response has been reported in TNBC. DOX is a cytotoxic anthracycline antibiotic that commonly generates resistance
[31][74]. The high expression of the circular RNA dubbed circUBE2D2 has been associated with a poor prognosis of TNBC patients in the advanced stages of the disease, as well as with lymph node metastases and resistance to DOX. CircUBE2D2 acts as a molecular sponge for miR-512-3p, which has a role as a suppressor of various tumors by acting on cell division cycle associated protein 3 (CDCA3)
[32][63]. On the other hand, the hsa_circ_0092276/miR-384/ATG7 axis promotes autophagy and DOX resistance through the overexpression of hsa_circ_0092276
[33][64].
Remarkably, circRNAs that perform the reverse function have been studied. For instance, the exogenous overexpression of circKDM4C has inhibited DOX resistance as well as proliferation and metastasis in breast cancer. CircKDM4C is a sponge for miR-548p, and the overexpression of this miRNA reverses the attenuation of malignant phenotypes
[34][65].
Several studies have corroborated the participation of circRNAs in the modulation of the proapoptotic and antiapoptotic proteins involved in resistance to chemotherapy. The overexpression of circAMOTL1 in TNBC has been related to significant increases in paclitaxel resistance
[35][75]. The role of circRNAs in apoptosis reduction, that is, modulating the expressions of AKT and proapoptotic factors BAX and BAK as well as the antiapoptotic BCL-2 protein, has been well-documented
[36][76].
79. Conclusions
CircRNAs are a type of ncRNA that has various regulatory roles in aggressive TNBC. Evidence has shown that they may greatly influence cell proliferation, migration, metastasis, apoptosis, and chemoresistance. CircRNAs have a dual role in metastasis, as they may act as oncogenes and tumor-suppressing genes through the regulation of microRNAs. Furthermore, circRNAs have been detected in exosomes in the serums of TNBC patients, representing potential biomarkers of disease, and participate in the response to anticancer drugs in TNBC.