Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Jessie Wu and Version 1 by Manuel F Varela.
A phospholipid membrane covers all living cells, forming an impenetrable barrier circumvented by solute transporters in the cell membrane. These proteins comprise energy-requiring systems, called active transporters, and those not requiring energy, called passive transporters. The major facilitator superfamily harbors thousands of transport proteins found in all living organisms, from bacteria to humans. Alignments of multiple amino acid sequences uncovered highly conserved sequence motifs are known to play important functional roles. One of these conserved sequences, the antiporter sequence motif or motif C, participates in the molecular mechanism of antimicrobial efflux in cancer cells and bacterial pathogens.
- antiporter motif
- major facilitator superfamily
- transporter
- antimicrobial resistance
- multidrug efflux
- bacteria
- cancer
1. Introduction
Bacterial physiology requires the availability of macromolecules and ions, as well as their precise balance concerning the external environment. The cell wall peptidoglycan provides the necessary stability to the cellular structure. In contrast, the cell membrane and its constituent proteins are critical in transporting solutes in and out of the cell in a coordinated manner. Although the transport process involves handling solutes as a major function, the implications of this function are more than the mere movements of substrates, as these processes are necessary for various other activities of bacteria involving metabolism, colonization, communication, virulence, and community living [1,2][1][2]. Transporter proteins are a large group of proteins that play critical roles in the physiology of bacteria by transporting essential macromolecules into the cell and extruding toxic metabolites, chemicals, and xenobiotics, maintaining cell homeostasis and helping bacteria survive in a wide range of environmental conditions. These proteins are embedded in the outer membrane of bacteria. They are a transportation conduit and an important means of communication with the external environment.
2. Transporter Biology
The transport proteins in living cells employ several different mechanisms to perform the activity and vary widely concerning their structures, types, and range of substrates, as well as the sources of energy that drive the transport process across the biological membrane [8,10][3][4]. The bacterial outer cell membrane functions as a protective barrier that selectively allows the movement of solutes across it into the periplasmic area. Since most solutes cannot cross the membrane barrier, specific transporters move substrates into and out of the bacterial cell. The simplest type of solute movement across the cell membrane occurs by passive diffusion of molecules such as certain gases (CO2 and O2) and water from a higher concentration to a lower concentration (downhill) without the involvement of transporter proteins. On the other hand, facilitated diffusion is enabled by carrier proteins that bind solutes and move them across the membrane through conformational changes. In contrast, channel proteins facilitate the movement of specific molecules through open pores formed by them [11][5]. As in passive diffusion, facilitated diffusion is not energy-coupled, although the concentration and the electrochemical gradient determine the direction of the movement of the substrate, and is always downhill (Figure 1).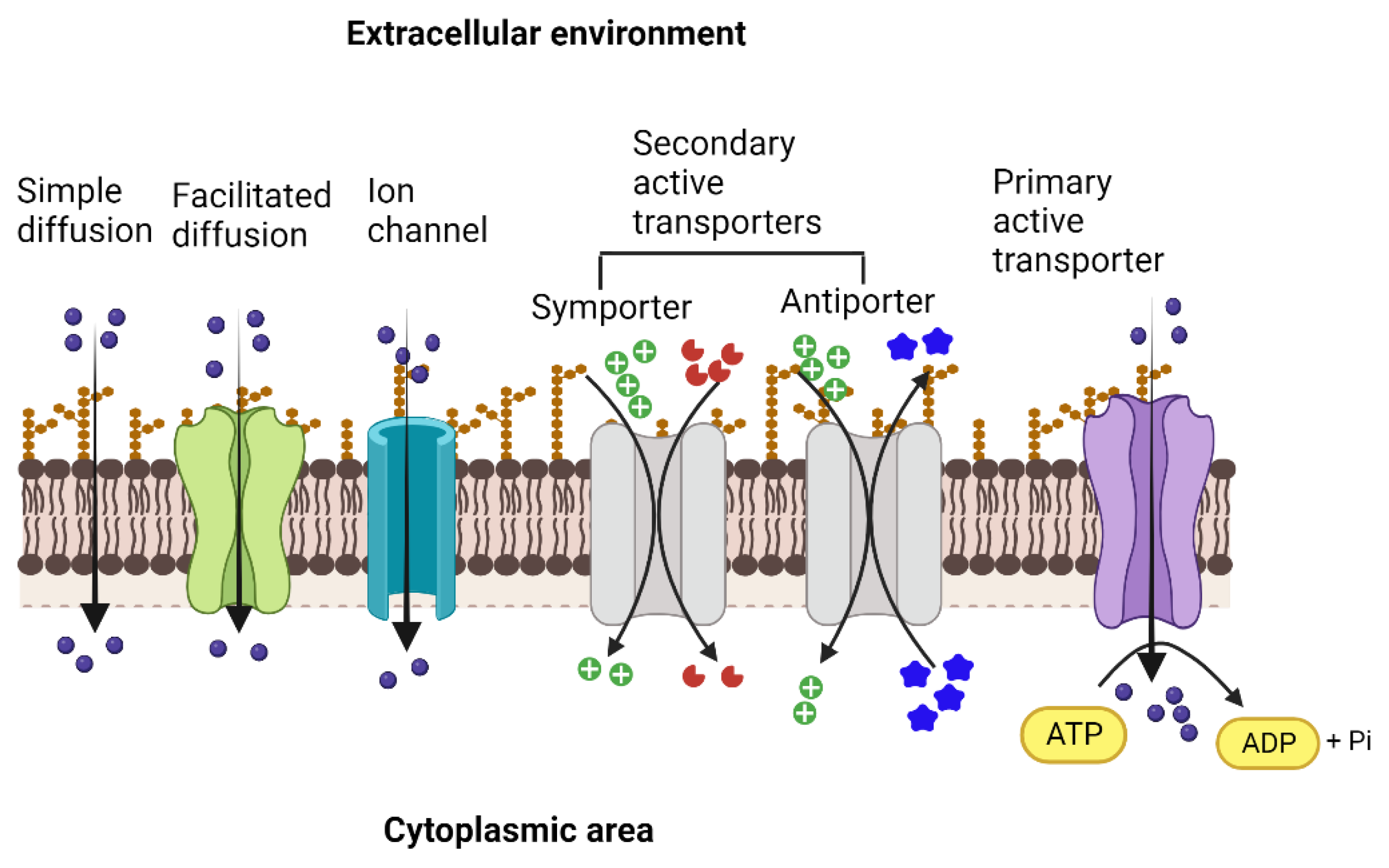
Figure 1. Types of membrane transporters that enable the movement of ions and solutes across the membrane into the cytoplasmic area and vice versa. Simple diffusion of gases and ions across the membrane facilitated diffusion and movement through ion channels (ungated or gated) that occur down the concentration gradient and are not coupled with energy sources. Both primary active and secondary active transporters move the solutes against the concentration gradient. They are energized either by the hydrolysis of ATP (primary active) or by the movement of ions, such as protons or sodium, driven by the electrochemical gradient across the membrane (secondary active). In the case of secondary active transport, the energetic driving force of one solute moving down its electrochemical gradient is coupled to the movement of the other solute moving up its concentration gradient. Created with BioRender.com.
3. Superfamilies of Transporters
Many transport proteins have been identified over the years. These solute transporters have diverse structures and functions. However, significant degrees of sequence identities and homologies are shared. Thus, a need for classifying these proteins akin to the Enzyme Commission (EC) system for enzymes, based on certain characteristics that distinguish them into distinct groups, was realized. This effort led to the creation of the transporter classification (TC) system (http://www.tcdb.org/ accessed on 24 September 2023), a curated database in which transporter proteins are systematically grouped based on specific characteristics, including the mode of transport, energy coupling mechanisms, sequence homology/protein phylogeny, topology, and substrate specificity [20,21,22,23][14][15][16][17]. The TC system follows the International Union of Biochemistry and Molecular Biology (IUBMB), an approved method of classification and nomenclature for transport proteins. Proteins originating from a common ancestor are homologous, share similar structures and functions, and are grouped into families or subfamilies. Accordingly, the database has over 1800 families of transport proteins grouped under distinct transporter classes, namely channels/pores, electrochemical potential-driven transporters, primary active transporters, group translocators, transmembrane electron carriers, auxiliary transport proteins, and transport protein families of unknown classification [23,24][17][18]. The secondary active transport proteins, i.e., symporters and antiporters, are grouped under the Electrochemical Potential-driven Transporters category and are distinct from the uniporters, which move solutes across the membrane down their gradients. The antiporter proteins transport two molecules simultaneously in opposite directions, energized by the proton-motive force gradient of H+ or Na+ across the plasma membrane, and are grouped under four superfamilies: (i) the major facilitator superfamily (MFS), (ii) the resistance-nodulation-cell division (RND) superfamily, (iii) the drug/metabolite transporter (DMT) superfamily, and (iv) the multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) superfamily [25][19] (Figure 2). The RND transporters are tripartite structures, forming multi-component complexes with an outer membrane channel and a periplasmic adaptor protein [26][20]. The multidrug and toxin extrusion (MATE) family of antimicrobial efflux pumps, which use both H+ and Na+ as energy sources, belong to the MOP superfamily [27,28][21][22]. Some of the well-characterized drug/Na+ antiporters include YdhE of Escherichia coli, NorM of Vibrio parahaemolyticus [29][23], NorM and VcmA of Vibrio cholerae [29[23][24],30], AbeM of Acinetobacter baumannii [31][25], and BexA of Bacteroides thetaiotaomicron [32][26]. The drug/H+ antiporters, such as the QacE and AbeS of Acinetobacter baumannii; QacC and SepA of S. aureus; EmrE, YnfA and MdtJ of E. coli and KpnEF of Klebsiella pneumoniae, belonging to the small multidrug resistance (SMR) family, are placed under the DMT superfamily [33,34,35][27][28][29].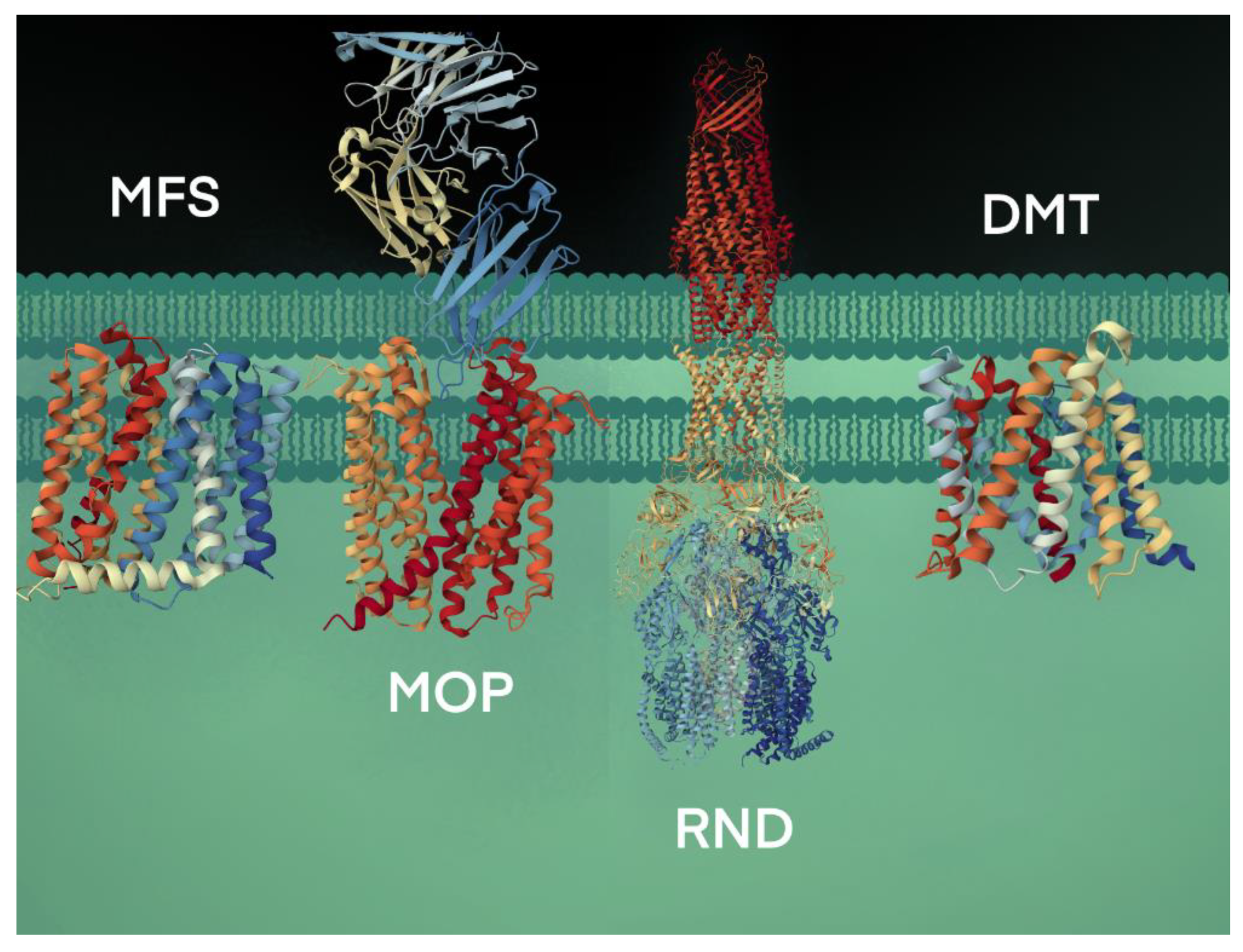
Figure 2. Bacterial efflux pumps of antiport type are grouped under four secondary active transporter superfamilies. The transport of substrates is coupled with H+ or Na+ ions. The representative MFS protein shown is the crystal structure of MdfA, a multidrug efflux pump (PBD code, 4ZOW) from E. coli [36][30]. The MOP protein shown is a high-resolution crystal structure using cryogenic electron microscopy analysis of NorM, a MATE transporter bound to a Fab molecule (PBD, 7PHP) from V. cholerae [37][31]. The RND transport system shown is the AcrAB-TolC crystal structure (PBD, 5V5S), a multipartite complex from E. coli that spans the inner (AcrB) and outer (TolC) membrane and periplasm (AcrA) [38][32]. The DMT crystal structure is the YddG transporter (PBD, 5I20) from the bacterium Starkeya novella [39][33].
References
- Hediger, M.A.; Romero, M.F.; Peng, J.B.; Rolfs, A.; Takanaga, H.; Bruford, E.A. The ABCs of solute carriers: Physiological, pathological and therapeutic implications of human membrane transport proteins. Introduction. Pflug. Arch. 2004, 447, 465–468.
- Davies, J.S.; Currie, M.J.; Wright, J.D.; Newton-Vesty, M.C.; North, R.A.; Mace, P.D.; Allison, J.R.; Dobson, R.C.J. Selective Nutrient Transport in Bacteria: Multicomponent Transporter Systems Reign Supreme. Front. Mol. Biosci. 2021, 8, 699222.
- Saier, M.H., Jr. Molecular phylogeny as a basis for the classification of transport proteins from bacteria, archaea and eukarya. Adv. Microb. Physiol. 1998, 40, 81–136.
- Konings, W.N.; Poolman, B.; van Veen, H.W. Solute transport and energy transduction in bacteria. Antonie Leeuwenhoek 1994, 65, 369–380.
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Carrier proteins and active membrane transport. In Molecular Biology of the Cell, 4th ed.; Garland Science: Oxford, UK, 2002.
- Tanaka, K.J.; Song, S.; Mason, K.; Pinkett, H.W. Selective substrate uptake: The role of ATP-binding cassette (ABC) importers in pathogenesis. Biochim. Biophys. Acta BBA Biomembr. 2018, 1860, 868–877.
- Akhtar, A.A.; Turner, D.P. The role of bacterial ATP-binding cassette (ABC) transporters in pathogenesis and virulence: Therapeutic and vaccine potential. Microb. Pathog. 2022, 171, 105734.
- Locher, K.P.; Lee, A.T.; Rees, D.C. The E. coli BtuCD structure: A framework for ABC transporter architecture and mechanism. Science 2002, 296, 1091–1098.
- Oldham, M.L.; Chen, J. Snapshots of the maltose transporter during ATP hydrolysis. Proc. Natl. Acad. Sci. USA 2011, 108, 15152–15156.
- Rismondo, J.; Schulz, L.M. Not just transporters: Alternative functions of ABC transporters in Bacillus subtilis and Listeria monocytogenes. Microorganisms 2021, 9, 163.
- Bilsing, F.L.; Anlauf, M.T.; Hachani, E.; Khosa, S.; Schmitt, L. ABC Transporters in bacterial nanomachineries. Int. J. Mol. Sci. 2023, 24, 6227.
- Hollenstein, K.; Dawson, R.J.; Locher, K.P. Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 2007, 17, 412–418.
- Okada, U.; Yamashita, E.; Neuberger, A.; Morimoto, M.; van Veen, H.W.; Murakami, S. Crystal structure of tripartite-type ABC transporter MacB from Acinetobacter baumannii. Nat. Commun. 2017, 8, 1336.
- Ren, Q.; Paulsen, I.T. Comparative analyses of fundamental differences in membrane transport capabilities in prokaryotes and eukaryotes. PLoS Comput. Biol. 2005, 1, e27.
- Saier, M.H., Jr.; Tran, C.V.; Barabote, R.D. TCDB: The Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006, 34 (Suppl. S1), D181–D186.
- Busch, W.; Saier, M.H. The IUBMB-endorsed transporter classification system. Mol. Biotechnol. 2004, 27, 253–262.
- Saier, M.H., Jr. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 2000, 64, 354–411.
- Saier, M.H.; Reddy, V.S.; Moreno-Hagelsieb, G.; Hendargo, K.J.; Zhang, Y.; Iddamsetty, V.; Lam, K.J.K.; Tian, N.; Russum, S.; Wang, J.; et al. The Transporter Classification Database (TCDB): 2021 update. Nucleic Acids Res. 2021, 49, D461–D467.
- Saier, M.H., Jr.; Reddy, V.S.; Tsu, B.V.; Ahmed, M.S.; Li, C.; Moreno-Hagelsieb, G. The Transporter Classification Database (TCDB): Recent advances. Nucleic Acids Res. 2016, 44, D372–D379.
- Zgurskaya, H.I.; Nikaido, H. Multidrug resistance mechanisms: Drug efflux across two membranes. Mol. Microbiol. 2000, 37, 219–225.
- Hvorup, R.N.; Winnen, B.; Chang, A.B.; Jiang, Y.; Zhou, X.F.; Saier, M.H., Jr. The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur. J. Biochem. 2003, 270, 799–813.
- Kuroda, T.; Tsuchiya, T. Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta BBA Proteins Proteom. 2009, 1794, 763–768.
- Morita, Y.; Kodama, K.; Shiota, S.; Mine, T.; Kataoka, A.; Mizushima, T.; Tsuchiya, T. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 1998, 42, 1778–1782.
- Huda, M.N.; Morita, Y.; Kuroda, T.; Mizushima, T.; Tsuchiya, T. Na+-driven multidrug efflux pump VcmA from Vibrio cholerae non-O1, a non-halophilic bacterium. FEMS Microbiol. Lett. 2001, 203, 235–239.
- Su, X.-Z.; Chen, J.; Mizushima, T.; Kuroda, T.; Tsuchiya, T. AbeM, an H+-coupled Acinetobacter baumannii multidrug efflux pump belonging to the MATE family of transporters. Antimicrob. Agents Chemother. 2005, 49, 4362–4364.
- Miyamae, S.; Ueda, O.; Yoshimura, F.; Hwang, J.; Tanaka, Y.; Nikaido, H. A MATE family multidrug efflux transporter pumps out fluoroquinolones in Bacteroides thetaiotaomicron. Antimicrob. Agents Chemother. 2001, 45, 3341–3346.
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 2018, 73, 2003–2020.
- Jack, D.L.; Yang, N.M.; Saier, M.H., Jr. The drug/metabolite transporter superfamily. Eur. J. Biochem. 2001, 268, 3620–3639.
- Huang, L.; Wu, C.; Gao, H.; Xu, C.; Dai, M.; Huang, L.; Hao, H.; Wang, X.; Cheng, G. Bacterial multidrug efflux pumps at the frontline of antimicrobial resistance: An overview. Antibiotics 2022, 11, 520.
- Heng, J.; Zhao, Y.; Liu, M.; Liu, Y.; Fan, J.; Wang, X.; Zhao, Y.; Zhang, X.C. Substrate-bound structure of the E. coli multidrug resistance transporter MdfA. Cell Res. 2015, 25, 1060–1073.
- Bloch, J.S.; Mukherjee, S.; Kowal, J.; Filippova, E.V.; Niederer, M.; Pardon, E.; Steyaert, J.; Kossiakoff, A.A.; Locher, K.P. Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins. Proc. Natl. Acad. Sci. USA 2021, 118, e2115435118.
- Wang, Z.; Fan, G.; Hryc, C.F.; Blaza, J.N.; Serysheva, I.I.; Schmid, M.F.; Chiu, W.; Luisi, B.F.; Du, D. An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. eLife 2017, 6, e24905.
- Tsuchiya, H.; Doki, S.; Takemoto, M.; Ikuta, T.; Higuchi, T.; Fukui, K.; Usuda, Y.; Tabuchi, E.; Nagatoishi, S.; Tsumoto, K.; et al. Structural basis for amino acid export by DMT superfamily transporter YddG. Nature 2016, 534, 417–420.
- Kumar, S.; Varela, M.F. Biochemistry of bacterial multidrug efflux pumps. Int. J. Mol. Sci. 2012, 13, 4484–4495.
- Pao, S.S.; Paulsen, I.T.; Saier, M.H., Jr. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998, 62, 1–34.
- Drew, D.; North, R.A.; Nagarathinam, K.; Tanabe, M. Structures and general transport mechanisms by the major facilitator superfamily (MFS). Chem. Rev. 2021, 121, 5289–5335.
- Yin, Y.; He, X.; Szewczyk, P.; Nguyen, T.; Chang, G. Structure of the multidrug transporter EmrD from Escherichia coli. Science 2006, 312, 741–744.
- Zhai, G.; Zhang, Z.; Dong, C. Mutagenesis and functional analysis of SotB: A multidrug transporter of the major facilitator superfamily from Escherichia coli. Front. Microbiol. 2022, 13, 1024639.
- Jiang, D.; Zhao, Y.; Wang, X.; Fan, J.; Heng, J.; Liu, X.; Feng, W.; Kang, X.; Huang, B.; Liu, J. Structure of the YajR transporter suggests a transport mechanism based on the conserved motif A. Proc. Natl. Acad. Sci. USA 2013, 110, 14664–14669.
- Nava, A.R.; Mauricio, N.; Sanca, A.J.; Domínguez, D.C. Evidence of calcium signaling and modulation of the LmrS multidrug resistant efflux pump activity by Ca2+ ions in S. aureus. Front. Microbiol. 2020, 11, 573388.
- Floyd, J.L.; Smith, K.P.; Kumar, S.H.; Floyd, J.T.; Varela, M.F. LmrS is a multidrug efflux pump of the major facilitator superfamily from Staphylococcus aureus. Antimicrob. Agents Chemother. 2010, 54, 5406–5412.
- Stephen, J.; Salam, F.; Lekshmi, M.; Kumar, S.H.; Varela, M.F. The major facilitator superfamily and antimicrobial resistance efflux pumps of the ESKAPEE pathogen Staphylococcus aureus. Antibiotics 2023, 12, 343.
- Crimmins, G.T.; Herskovits, A.A.; Rehder, K.; Sivick, K.E.; Lauer, P.; Dubensky, T.W., Jr.; Portnoy, D.A. Listeria monocytogenes multidrug resistance transporters activate a cytosolic surveillance pathway of innate immunity. Proc. Natl. Acad. Sci. USA 2008, 105, 10191–10196.
- Sharma, A.; Gupta, V.K.; Pathania, R. Efflux pump inhibitors for bacterial pathogens: From bench to bedside. Indian. J. Med. Res. 2019, 149, 129.
- Soto, S.M. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 2013, 4, 223–229.
More
