The intracellular concentrations of oxygen and reactive oxygen species (ROS) in living cells represent critical information for investigating physiological and pathological conditions. Real-time measurement often relies on genetically encoded proteins that are responsive to fluctuations in either oxygen or ROS concentrations. The direct binding or chemical reactions that occur in their presence either directly alter the fluorescence properties of the binding protein or alter the fluorescence properties of fusion partners, mostly consisting of variants of the green fluorescent protein. Oxygen sensing takes advantage of several mechanisms, including (i) the oxygen-dependent hydroxylation of a domain of the hypoxia-inducible factor-1, which, in turn, promotes its cellular degradation along with fluorescent fusion partners; (ii) the naturally oxygen-dependent maturation of the fluorophore of green fluorescent protein variants; and (iii) direct oxygen binding by proteins, including heme proteins, expressed in fusion with fluorescent partners, resulting in changes in fluorescence due to conformational alterations or fluorescence resonance energy transfer.
- oxygen sensor
- reactive oxygen species
- fluorescent proteins
1. Relevance of O2 and ROS Sensing
2. Biosensors for O2
2.1. O
2
Sensing Mediated by Prolyl Hydroxylases
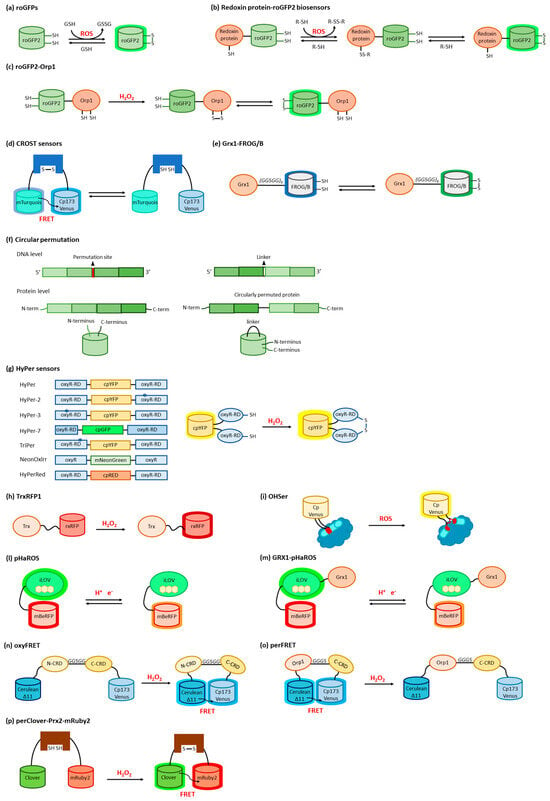
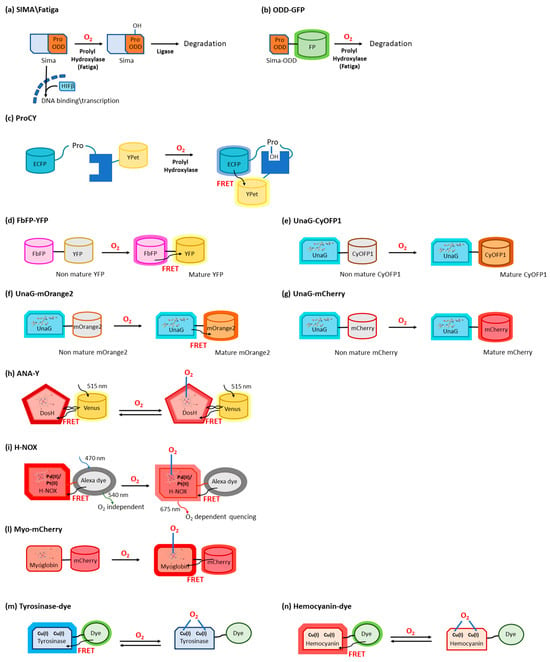
The transcription factor hypoxia-inducible factor-1 (HIF-1) is the main actor in modulating O2-mediated gene expression levels in all Parahoxozoa. HIF-1 is a αβ heterodimer with the levels of the β subunit being O2-independent, whereas those of the α subunit are O2-dependent [3][10] through its O2-dependent proteolysis [4][11]. In fact, the α subunit is constitutively synthesized but rapidly degraded under normoxic conditions [5][12] as a consequence of enzymatic hydroxylation of conserved proline residues (Pro402 and Pro564) located in its oxygen-dependent degradation domain (ODD) [6][7][13,14]. The enzymes responsible for proteolysis are 2-oxoglutarate-dependent dioxygenases containing a prolyl hydroxylase domain (PHD) [8][15]. Hydroxylated HIF-1α binds to the von Hippel–Lindau tumor suppressor protein (VHL) and is then ubiquitylated by the VHL-E3 ligase complex (Figure 1a).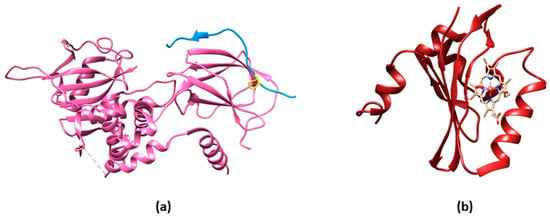
2.2. O
2
Sensors Based on O
2
-Dependent Maturation of GFP Variants
2.3. O
2
Sensors Based on O
2
-Binding Heme Proteins
Oxygen sensing has been also approached by exploiting heme-binding proteins, which use the iron-containing heme group as the O2 sensing element to be transduced in other cellular signals. These constructs are commonly composed of an N-terminal heme-containing globin domain, whose conformational variation upon O2 binding activates a functional domain that triggers a catalytic domain. This latter can have a diguanylate cyclase (DGC) [26][37] or phosphodiesterase (PDE) [27][38] activity toward cyclic diGMP (c-diGMP) and histidine kinase (HK) activity [28][39]. An example is a genetically encoded O2 biosensor based on the direct O2 sensor DosP from E. coli. This protein contains a heme-binding globin domain called DosH (Figure 1b) and a PDE catalytic domain, which converts cyclic-di-GMP to linear-di-GMP [29][40]. However, based on the crystallographic structures [10][30][18,41], the conformational variation of DosP upon O2 binding or dissociation is not sufficient to develop a suitable FRET signal; to overcome this limitation, DosH has been associated with a fluorescent protein to exploit the heme spectroscopic changes in Soret and Q peaks upon O2 binding shifting from 425 and 560 nm to 414 and 580 nm, respectively [31][42]. The yellow FP Venus—a GFP variant with a suitable spectral overlap with DosH—was conjugated with DosH using an optimized antiparallel coiled-coil linker. When Venus was excited at 500 nm, its fluorescence emission at 527 nm was absorbed by DosH in the O2-free form, resulting in low Venus quantum yield; when DosH was O2-bound, Venus’s fluorescence intensity increased. The change in heme absorption was therefore amplified by a change in fluorescence intensity. This sensor, called ANA-Y (anaerobic/aerobic sensor yellow), was used to sense O2 in the micromolar range (Figure 2h) [32][43]. The heme nitric oxide/oxygen-binding protein (H-NOX) of the thermophilic bacterium Caldanaerobacter subterraneus has been exploited to develop an O2 sensor as a robust protein scaffold also able to bind unnatural heme cofactors. The natural heme was replaced with a Pd(II) or Pt(II) porphyrin, a phosphorescent cofactor (650–800 nm range) that, in the triplet excited state, can interact with molecular oxygen; in particular, Pd(II) porphyrins are more sensitive to low O2 levels and Pd(II) has a larger range of O2 sensitivity. A ratiometric sensor based on H-NOX incorporating Pd(II) or Pt(II) porphyrins was developed by conjugating to the protein an Alexa fluorescent dye that is the O2-independent FRET donor to the porphyrin. The selective excitation of Alexa dye guarantees that the porphyrin emission will only be from FRET, thus minimizing the background signal (Figure 2i) [33][44]. As a different approach, the spectroscopic changes of heme upon O2 binding have been exploited and amplified in a fusion protein (Myo-mCherry), where myoglobin has been fused with mCherry fluorescent protein; spectral changes resulted in a change of FRET, captured as a change in fluorescence lifetime within cells by FLIM (Figure 2l).2.4. O
2
Sensors Based on O
2
-Binding Copper Proteins
The use of type 3 copper proteins to sense O2 has been also approached; these proteins have a binding site with two Cu(I) ions that specifically bind to O2, and the complex, once formed, gives absorption peaks at 340 and 570 nm [34][47]. O2 binding typically also causes Trp fluorescence changes, but the use of fluorescence coming from an exogenous fluorophore with a more efficient and higher-wavelength emission is made necessary to increase sensitivity (Trp has a moderate quantum yield, around 0.14) and to allow the detection in tissues showing significant autofluorescence. An example is given by the tyrosinase from the bacterium Streptomyces antibioticus and hemocyanin from the arthropod Carcinus aestuarii, which have been conjugated at the N-terminus with different fluorescent dyes (Alexa 350, Atto390, Cy3, Cy5, Atto655), showing an absorption overlapping Trp emission, thus allowing the detection as a variation in FRET upon O2 binding (Figure 2m) [35][48]. The detection of a single O2 molecule has been obtained by exploiting the O2 carrier hemocyanin from the tarantula Eurypelma californicum. This protein shows spectral differences when it binds oxygen, and these absorption changes can be used to sense O2 presence. Moreover, to amplify the signal, TAMRA fluorescent dye was conjugated with hemocyanin, and its fluorescence quantum yield was shown to decrease by a factor of two when hemocyanin was oxygenated [36][49]. This was caused by a FRET where TAMRA was the donor and the O2 binding site was the acceptor (Figure 2n) [37][50].3. Biosensors for ROS
3.1. ROS Sensors Based on roFPs
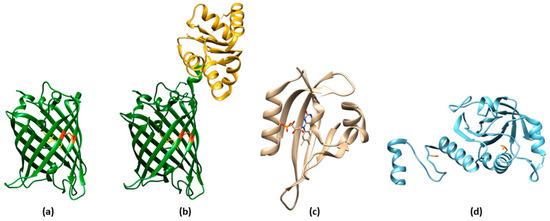
Reduction-oxidation sensitive GFPs (roGFPs) are non-natural variants of A. victoria GFP obtained by substituting surface-exposed residues with cysteine residues appropriately distanced to form disulfide bonds at suitable redox potentials (Figure 3a,b and Figure 4a). The first examples of roFPs consisted of the introduction of an artificial pair of cysteine residues on YFP, giving rise to one of the first classes of redox-sensitive protein-based fluorescent sensors. The first rxYFP was conceived in 2001 by Winther’s research group, when the YFP sequence was modified to mutate Asn149 and Ser202 to two cysteine residues able to form a disulfide bridge under oxidizing conditions (Figure 3a) [38][51].