2.1. Utility and Accuracy of EUS-nCLE
EUS-nCLE provides real-time microscopic analysis of PCL epithelium without the need for high-risk endoscopic biopsy or surgical excision
[18][19][43,44]. The PCL is visualized using EUS, and intravenous fluorescein is injected 2–3 minutes prior to imaging. A 19-gauge FNA needle preloaded with an nCLE miniprobe is advanced into the cyst until it opposes the cyst epithelium. The miniprobe is moved throughout the cyst cavity to assess different areas of its internal lining for approximately 6 minutes. After the video sequence is acquired, the miniprobe is removed, and the cyst fluid is aspirated for further analysis
[20][45].
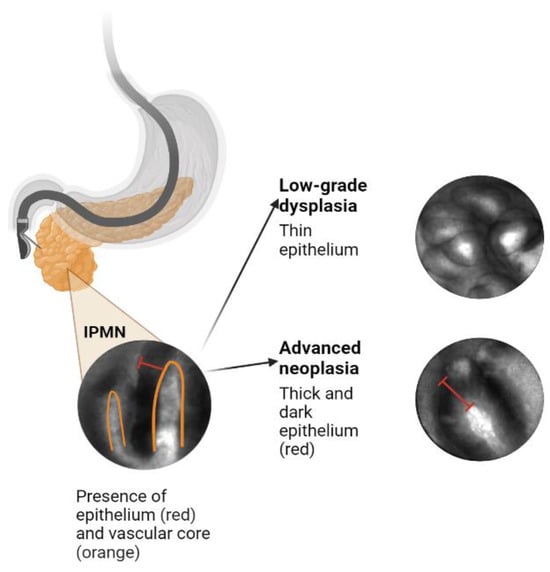
Several features that can be visualized on EUS-nCLE, which correspond to a higher histologic grade. For example, papillary epithelial width, as measured on nCLE images, suggests cellular atypia, while increased epithelial darkness on images is associated with nuclear stratification. These characteristics demonstrated high sensitivity in predicting HGD-Ca in an analysis of 26 BD-IPMNs. Papillary epithelial “width” and “darkness” exhibited sensitivities of 90% and 91%, respectively
[21][48]. Surgical histopathology served as the reference standard in this analysis. Additionally, there was substantial interobserver agreement (IOA) among external nCLE experts in detecting HGD-Ca, with a κ value of 0.61 for epithelial width and 0.55 for darkness
[21][48].
2.2. CNN-AI Algorithm for nCLE Analysis
By employing accurate AI-driven image analysis and recognition, the need for time-consuming manual quantification of papillary epithelial parameters by endomicroscopy specialists can be bypassed. In a recent post hoc analysis of video frames from the INDEX study, which included patients with histopathologically proven IPMNs, two CNN-based algorithms were developed to detect HGD-Ca in the lesions
[20][45]. The first algorithm utilizes an instance-segmentation-based model, specifically Mask R-CNN, to detect and segment papillary structures. It then measures papillary epithelial thickness and darkness and employs these features for diagnosing HGD-Ca. This model achieved an accuracy of 82.9%. The second algorithm applies a CNN model, namely VGGNet, to directly extract features from the holistic nCLE video frames for risk stratification. This approach yielded an accuracy of 85.7%. In comparison, the accuracy of current society guidelines (AGA and Fukuoka) reached only 68.6% and 74.3%, respectively. These findings highlight the potential of AI-based approaches in improving diagnostic accuracy and outperforming existing guidelines in the assessment of IPMNs.
2.3. Improving and Prospectively Evaluating the Single-Center Algorithm
3.3. Improving and Prospectively Evaluating the Single-Center Algorithm
In addition to high accuracy, the single-center algorithm must also demonstrate the ability to incorporate new patient data. Many patients with PCLs require EUS with or without nCLE, with big data being stored as large video files. The mean duration of the unedited EUS-nCLE videos in previous studies was approximately seven minutes, and the videos were manually shortened to under three minutes of high-yield portions
[20][21][45,48]. Both algorithms in the pilot study used pre-edited videos where frames with artifact, blurring, or redundancy were removed by an nCLE expert. An algorithm that can be directly applied to unedited videos, without the need for manual editing, will greatly increase model efficiency, applicability, and generalizability.
The future plans involve the development of a video summarization AI algorithm that will convert unedited nCLE videos into shorter, high-yield video clips, which will, in turn, improve the performance of the CNN-based algorithm
[22][23][24][49,50,51]. This approach aims to streamline the diagnostic process for HGD-Ca detection in three steps. First, a CNN-based classifier will be used to classify edited video frames into “high-risk” and “low-risk” images based on the presence or absence of papillary structures (as an indicator of potential dysplasia)
[25][52]. Second, an instance-segmentation-based algorithm will be employed to segment papillae and measure papillary epithelial thickness and darkness, enabling the grading of dysplasia. Concurrently, a holistic-based algorithm will utilize a CNN-based model to extract features from nCLE images, focusing on identifying HGD-Ca. The outputs of these two algorithms will be appropriately fused to leverage their complementary abilities and reduce uncertainty in the diagnostic process. Finally, image-level predictions from the entire edited video will be aggregated using a majority voting approach to diagnose HGD-Ca in each subject.
2.4. Creating an Integrative Predictive Algorithm
While EUS-nCLE provides valuable information for PCL diagnosis, it represents only one source of patient data. Multiple clinical, demographic, genomic, and radiographic data points have been identified as significant predictors of HGD-Ca; incorporating all of these data into an integrative predictive algorithm will likely significantly improve accuracy. It is important to acknowledge that existing predictive models and expert consensus-led guidelines may oversimplify the impact of each variable considered. In routine medical practice, clinicians take into account patient demographics, apply guidelines such as Fukuoka-ICG, and often engage in multidisciplinary team discussions to assess the risk of malignancy associated with PCLs
[13].
Molecular analysis using NGS so far has been shown to more reliably predict HGD-Ca in IPMNs as compared to the standard of care. In a 2015 composite analysis, the combination of genetic mutations in SMAD4, RNF43, TP53, and aneuploidy predicted HGD-Ca in IPMNs with a sensitivity of 75% and a specificity of 92%
[25][52]. Another study showed that the presence of KRAS/GNAS along with additional mutations in TP53, PIK3CA, and PTEN produced 88% sensitivity and 97% specificity for BD-IPMNs with HGD-Ca
[26][53].
Standard-of-care variables include clinical characteristics (age, gender, onset of diabetes, family history symptoms, pancreatitis history), serum CA 19-9, cyst fluid analysis (glucose, CEA, cytology) as well as cyst and pancreas morphology, as detected via CT/MRI/EUS (factors, such as size, wall, thickness, mural nodules, growth rate, and pancreatic duct diameter).
The second component aims to develop an ML-based approach for integrating the four data sources. Decision trees are chosen as the ML algorithm of choice due to their ability to handle both continuous and categorical variables seamlessly. Additionally, decision trees provide human-understandable explanations of the decision-making process, enabling experts to verify and interpret the learned decision trees. This feature facilitates the integration of the model into future management guidelines. Suitable options for doing this include using random forests and XGBoost, both of which are well-known ensemble methods over decisions trees that offer easy ways to control the model complexity to overcome over/under-fitting by controlling the number of input variables, tree depths/widths, and number of trees
[27][28][29][54,55,56]. These enhanced models have the potential to diagnose HGD-Ca in BD-IPMNs with optimized diagnostic accuracy. Furthermore, by identifying the most significant contributors from the data sources, these models can potentially inform new clinical practice guidelines and improve the risk stratification of BD-IPMNs.
2.5. Prior Integrative Algorithms
Integration of multiple data sources into AI algorithms to solve clinical problems related to PCLs and pancreatic malignancy have shown promising results
[25][30][31][32][52,57,58,59]. In 2015, Springer et al. developed an algorithm that analyzed multi-parametric features (known as Multivariate Organization of Combinatorial Alterations or MOCA) to identify PCLs requiring resection
[25][52]. The researchers combined composite clinical markers (such as age, presence of abdominal symptoms, and specific imaging results) and composite molecular markers (including aneuploidy and various gene mutations) in their MOCA algorithm. When evaluating the markers individually, the composite clinical marker and the composite molecular marker showed sensitivities of 75% and 77%, respectively, in identifying cysts that required resection. However, when used together, their sensitivity increased to 89%. Furthermore, the combination of molecular and clinical markers in the study resulted in a sensitivity of 94% for detecting IPMNs, an increase from the 76% sensitivity observed with the composite molecular marker alone. Subsequently, the same group developed CompCyst in 2019, which is a test built on the MOCA algorithm. This test categorizes patients with PCLs into three management groups (surgery, surveillance, or discharge) based on the evaluation of various clinical and molecular markers
[31][58]. Overall, these advancements in combining molecular and clinical markers, as demonstrated by the MOCA algorithm and CompCyst, offer improved stratification and management of patients with PCLs. The test integrated three data elements: presence of VHL mutation but absence of GNAS mutation, decreased expression of a VEGF-A protein, and a combination of factors (solid component of cyst seen on imaging, aneuploidy, and presence of mutations in certain genes). Clinical, imaging, and molecular data were integrated in their test, and they produced higher accuracy compared to conventional clinical and imaging criteria. The authors estimated that widespread use of CompCyst may reduce the number of unnecessary surgical resections by 60%.
Other recent studies have also leveraged ML to perform integrative analyses to manage other malignancies. One study used a deep-learning-based stacked ensemble model to predict the prognosis of breast cancer from available multi-modal cancer datasets, including genetic data (gene expression, copy number variation) and clinical data (age, subjective timing of menstruation and menopause, timing of pregnancy and others)
[33][64]. Integrative approaches have also been applied to determine prognosis of clear cell renal cell carcinoma and lung adenocarcinoma
[34][35][65,66].