Ulcerative colitis and Crohn’s disease are traditionally defined as the two main subtypes of inflammatory bowel disease. However, a more recent view considers inflammatory bowel diseases (IBDs) as a spectrum of heterogeneous phenotypes with consistent differences in clinical presentation and behaviors, likely explained by differences in underlying pathogenetic mechanisms. The etiology is still elusive, and the suggested pathogenesis is a complex interplay among genetic predisposition and abnormal immune response at the mucosal intestinal level, activated by only partially identified environmental triggers leading to altered intestinal permeability and impaired handling of gut microbiota. The undeniable continuous progress of medical therapy with more frequent shifts from traditional to more advanced modalities also underlines the actual unmet needs.
1. Introduction
Inflammatory bowel diseases (IBDs) are progressively increasing their prevalence worldwide with over 2 million North Americans and 2 million Europeans currently diagnosed. Although the incidence has almost plateaued in these countries, it is steadily increasing elsewhere with probably 6 million affected globally
[1]. Therefore, a progressive increase in the burden for society with massive pressure on health care costs worldwide is expected. The clinical variability of behaviors is striking with cases with limited involvement and a favorable course and probably the majority with progressive bowel damage and complications leading to hospitalization, surgery and eventually cancer
[2][3][2,3]. In addition, up to half of patients may have extraintestinal manifestation, and as a whole, all have an increased risk for other immune-mediated diseases. It is frustrating to consider that still the possibility to prognosticate the disease course is based on rather few and yet not solid and accurate clinical features, lacking appropriate biomarkers.
Although the therapeutic armamentarium is continuously expanding, approximately one-third of the patients are primary non-responders to the initial treatment, and up to half will have incomplete or a loss of response over time.
2. The Potential of Precision Medicine IBD
The concept of precision medicine is not new and was first reported in a publication in 1971
[4][7]. However, since then, many other terminologies have been used and eventually not with the same meaning, such as “individualized medicine”, “personalized medicine”, “tailored medicine”, etc. Of note, the US National Council Committee
[5][8] stated that the term “precision medicine” should be used for the “
tailoring of medical treatment to the individual characteristics of each patient to classify individual in subpopulations that differ in their susceptibility to a particular disease or their response to a specific treatment”. In addition, the Committee clearly stated the distinction between “precise” and “personalized” medicine by clarifying that “
personalized medicine refers to treatment tailored towards single individual” while “
precision medicine seeks to identify homogenous subgroups stratified according to a similar behavior”.
More recently, another publication
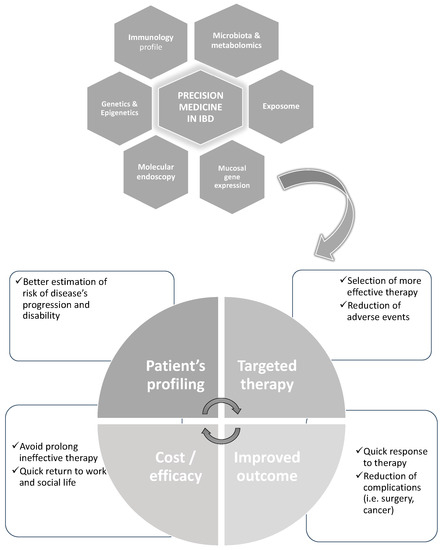
[6][9] provided a more extensive and modern view stating that “PM seeks to improve stratification and timing of health care by utilizing biological information and biomarkers at the level of molecular disease pathways, genetics, proteomics as well as metabolomics”—essentially, embracing the concept of the application of system biology and bioinformatics in health care. System biology allows the possibility of analyzing the interaction of all the components in a well-defined biological context
[7][10]. In the scenario of IBD, this means analyzing the role of environmental factors (so-called exposome), predisposing genetic factors (genome), the possible non-genetic modification of the genome (epigenome) and gene expression at the mucosal levels including coding and non-coding RNAs (transcriptome and proteome), as well as the differences in gut microbiota and their produced metabolites (microbiome and metabolome), all at once (
Figure 1).
Figure 1. An example of the potential of the evaluation by system biology of multi-omics in IBD aiming to achieve with more “precision” a tailored therapy and better outcomes.
3. The Potential of Artificial Intelligence in IBD
Any attempt for a single investigator to analyze all the possible information related to the “omics” of a single patient with IBD, resulting in millions of data points, is obviously grossly inadequate. Rather, AI and its computational revolution and speed may support and unravel the underlying complexity of such biology. This may likely be achieved with the use of ML, essentially, a branch of AI in which computer algorithms improve automatically through the experience, in a hypothesis-free context
[8][12].
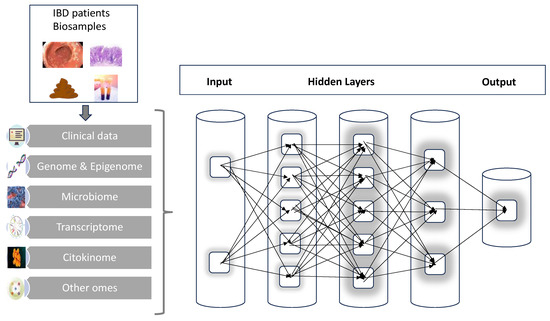
Conceptually, a possible structure for an IBD neural network can be organized into three major layers. The first layer is where all the input is conveyed, responsible for handling the individual IBD omics such as genome, transcriptome, etc. Next, it must imagine hidden layers that will perform all the complex calculations, interactions and combinations of parameters. Finally, there will be the output layer that, after receiving the signals for the hidden layers, will finally process and provide the results (
Figure 2). There are already preliminary applications and results of this methodology that will be detailed in the next sections. There is, however, still the need to evaluate in IBD which is the best computation tool for the specific information that is under investigation.
Figure 2. A graphic scheme of the multi-step process of analysis starting from patient’s biosamples; the combination of clinical data and multi-omics results in artificial neural networks will progressively elaborate the large complexity and number of data to deliver hopefully more individualized and precise output.
4. Initial Applications in IBD
4.1. Prediction of Disease Course
Given the great heterogeneity, it is very likely that understanding the disease course and evolution is one of the cornerstones for developing a precision medicine approach to IBD.
An emerging area is the comprehensive evaluation of intestinal involvement in CD not only with endoscopy but utilizing imaging-based biomarkers. More specifically, one potential tool is the so-called Lemann Index (LI) which is a scoring system that combines the utilization of clinical and endoscopic figures with imaging obtained with magnetic resonance enterography (MRE). The evaluation of bowel damage already at the diagnosis might help decision making in favor of more aggressive therapy, and periodic re-evaluation might estimate the increase or reduced disease burden [9][13].
Another important contribution has been achieved by the pediatric cohort of the RISK study [10][15] which has demonstrated that combining the clinical information (age, race, disease location), antimicrobial serologies and gene signature from ileal biopsies (more specifically, an extracellular matrix signature) with a system biology methodology has been able to predict the more aggressive course of the disease such as the development of stricturing disease in 3 years of disease course.
Because of the invasiveness of the endoscopic procedure and the restriction for more frequent re-evaluation, the focus has been shifted to the utilization of blood-based biomarkers. Unfortunately, neither the investigation of DNA methylation [11][16] nor the genomewide association studies (GWAs) [12][17] have demonstrated adequate power to identify variants with an elevated odds ratio (OR) which is useful to direct patient stratification.
4.2. Prediction of Complication of Therapy
Genotyping of the thiopurine S-methyltransferase (
TMPT) gene has been largely used to predict the possible bone marrow toxicity of thiopurines, determine the correct dosage
[13][26] and reduce toxicity. More recently, the polymorphism of the
NUDT15 gene has been identified as a major risk factor for the bone marrow toxicity of thiopurines
[14][27]. The combined polymorphism of these two genes explained almost 50% of the thiopurines-induced myelotoxicity. The polymorphism of the HLA-DQA1*02:01-HLA-DRB1*07:01 haplotype
[15][28] has been identified as predictive of thiopurine-induced pancreatitis, but given the rarity (number needed to test = 76), testing is limited in clinical practice.
4.3. Prediction of Response to Therapy
Given the substantial rate of primary non-response of any therapy for IBD as well as a lack of complete response or a loss of response, the possibility to predict the efficacy of the therapy and hence more precise tailoring of the management for every patient is probably the most important unmet need.
One fascinating option is the so-called “
molecular endoscopy”. Essentially, during the endoscopy, an anti-TNF fluorescent antibody liquid can be sprayed. Subsequently, with confocal laser endoscopy, it is possible to quantify the cells showing mTNF. By using this methodology, Atreya et al.
[16][35] demonstrated a 92% response to anti-TNF therapy after 3 months in a patient with CD with elevated mTNF expression; in contrast, the response to the therapy was only 13% in a patient with low mTNF expression.
The evaluation of
genetic polymorphisms in IBD has been particularly successful in identifying over 250 susceptibility markers of disease risk, but it is not very informative toward the prediction of the efficacy of therapy. More data have been obtained but often discordant about the response to anti-TNF therapy.
In IBD, there is definitely a change in the microbiota with reduced diversity and different abundances of some species; whether it is a primary or secondary phenomenon is still unclear
[17][24]. What is intriguing is the concept that differences in microbiota might also influence the response to the therapy
[18][41].
Conceptually, as earlier mentioned, the combination of more biomarkers with more sophisticated statistics could be in the future more productive. Some experiences are already accumulating. Barber et al.
[19][52], by utilizing a genetic score based on the Illumina Immunochip, did predict with higher accuracy compared to clinical parameters the response to anti-TNF. In addition, 16 polymorphisms did predict a more prolonged efficacy of the therapy. Similarly in the previously mentioned RISK study
[10][15], the combination of the information about fecal microbiota, the genetic profile and the presence of anti-microbic antibodies not only predicted a different course of the disease but also a better response to infliximab in some patient clusters.
More information will be also achieved in the future from ongoing prospective studies such as the trial PROFILE (ISRCTN11808228) using a top-down vs. step-up approach of therapy based on the immunologic profile
[20][23], as well as the study PREDICCT (ISRCNT67248113)
[21][54] looking at the “prognostic effects of environmental factors in Crohn’s and Colitis” that will monitor the activity and outcome of the disease, taking into consideration also the diet, lifestyle and microbiota.