DEAD-box开螟酶 decapping enzyme 20(DDX20)是一种假定的RNA开荬酶,可以通过保守基序Asp-Glu-Ala-Asp(DEAD)进行鉴定。细胞过程涉及许多RNA二级结构改变,包括翻译起始,核和线粒体剪接以及核糖体和剪接体的组装。据报道,DDX20在细胞转录和转录后修饰中起重要作用。一方面,DDX20可以与各种转录因子相互作用并抑制转录过程。另一方面,DDX20形成存活运动神经元复合体并参与snRNP的组装,最终影响RNA剪接过程。最后,DDX20可以潜在地依靠其RNA解开酶功能参与 (DDX20) is a putative RNA-decapping enzyme that can be identified by the conserved motif Asp–Glu–Ala–Asp (DEAD). Cellular processes involve numerous RNA secondary structure alterations, including translation initiation, nuclear and mitochondrial splicing, and assembly of ribosomes and spliceosomes. DDX20 reportedly plays an important role in cellular transcription and post-transcriptional modifications. On the one hand, DDX20 can interact with various transcription factors and repress the transcriptional process. On the other hand, DDX20 forms the survival motor neuron complex and participates in the assembly of snRNP, ultimately affecting the RNA splicing process. Finally, DDX20 can potentially rely on its RNA-unwinding enzyme function to participate in microRNA(miRNA)成熟,并作为RNA诱导的沉默复合物的组分。 (miRNA) maturation and act as a component of the RNA-induced silencing complex.
- DDX20
- NF-κB
- transcription
- SMN complexe
- cancer
- miRNA
1. 简介Introduction
2. Distribution, Structure, and Subcellular Localization of DDX20的分布、结构和亚细胞定位
作为具有ATP依赖性RNA解绕酶的s a novel human member of the DEAD-box家族的新型人类成员,DDX20在哺乳动物细胞的分子水平上可检测到,针对DDX20的单克隆抗体已升高至 family with ATP-dependent RNA-unwinding enzymes, DDX20 was detectable at the molecular level in mammalian cells with monoclonal antibodies against DDX20 elevated to 103 kDa[6]。随后的研究表明,编码DDX46蛋白的隐杆菌基因 in size [6]. Subsequent studies revealed that the Cryptobacterium hidradenum gene Mel-46 encoding the DDX20在其整个发育过程中表达,并在发育过程中发挥促进作用[7]。细胞水平的监测结果显示,在肝细胞癌组织中可检测到 protein is expressed throughout its development and plays a facilitating role during development [7]. The monitoring results at the cellular level revealed that decreased DDX20表达降低,从而影响疾病进程[8,9]。此外,在组织和器官水平上, expression could be detected in cancerous tissues of hepatocytes, thus affecting the disease process [8][9]. In addition, at the tissue and organ level, DDX20在睾丸中表达,甚至在产生类固醇和非类固醇的组织中表达[10]。此外, was expressed in the testis and even in steroid- and nonsteroid-producing tissues [10]. Furthermore, DDX20在某些癌组织中显著过表达,例如肝细胞癌和结直肠癌、前列腺癌和胃癌,通常表明预后良好[11,12,13,14]。 is significantly overexpressed in certain cancerous tissues, such as hepatocellular carcinoma and colorectal, prostate, and gastric cancers, and usually indicates a good prognosis [11][12][13][14]. DDX20首先由单性生殖多细胞生物 was first encoded by the genome of the parthenogenetic multicellular organism Dictyostelium discoideum的基因组编码,并保留在出生后动物中[15]。一些药物,如他汀类药物,也可以通过甲羟戊酸途径和R and is retained in postnatal animals [15]. Some drugs, such as statins, can alsoA下游抑制DDX20的表达[16]。 inhibit DDX20 expression through the mevalonate pathway and downstream of RhoA [16]. The DEAD-box RNA消解酶家族具有独特的Asp-Glu-Ala-Asp(-decapping enzyme family features a unique Asp–Glu–Ala–Asp (D-E-A-D)序列基序[17]。所有) sequence motif [17]. All DEAD-box开盖酶(包括 decapping enzymes, including DDX20)在N端都有一个保守的核心结构域,主要包括两个重组酶A(RecA)样结构域[18]。如图, possess a conserved core structural domain at the N-terminal end, primarily comprising two recombinase A (RecA)-like structural domains [18]. As shown in Figure 1所示,该核心结构域包括1个保守基序,即, this core structural domain includes nine conserved motifs, namely Q、I、Ia、Ib、II、III、IV、V和VI,它们参与ATP结合和水解、RNA底物结合、通过ATP结合水解调节开盖酶活性以及调节ATP酶活性[19]。, I, Ia, Ib, II, III, IV, V, and VI, which are involved in ATP binding and hydrolysis, RNA substrate binding, regulating decapping enzyme activity via ATP-binding hydrolysis, and regulating ATPase activity [1]. The D-E-A-D sequence motif is primarily found in 序列基序主要存在于高度保守的基序 II 中,这也是the highly conserved motif II, which is also the source of the “DEAD-box”名称的来源。非保守的N端和 name. Nonconserved N- and C-terminal auxiliary domains flank the core domain, ranging in size from a few to several hundred amino acids. These are mainly associated with specific functions of the decapping enzyme [19]. For instance, the C端辅助结构域位于核心结构域的两侧,大小从几个到几百个氨基酸不等。这些主要与开胃酶的特定功能有关[20]。例如,-terminal region of DDX20的C端区域是维持其解偶联解旋酶活性所必需的[<>]。 is necessary for it to maintain its uncoupling helicase activity [20].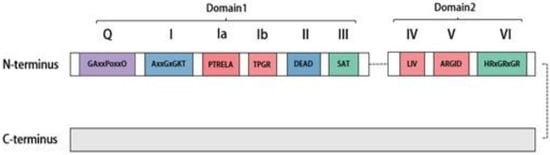
3. Splicing Features of DDX20的拼接特点
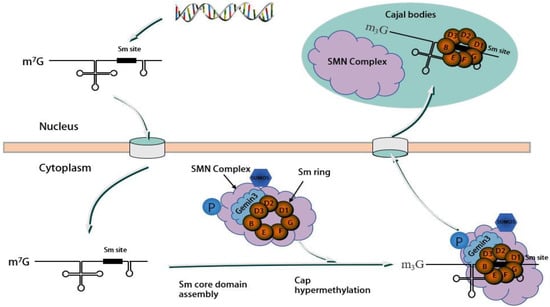
现有研究表明,运动神经元蛋白Existing research indicates that the motor neuron protein SMN与, along with Gemin2-8蛋白(包括–8 proteins, including DDX20/Gemin3和Unrip)一起形成稳定的SMN复合物,其中 and Unrip, form a stable SMN complex, with DDX20/Gemin3在该复合物的组装中起着至关重要的作用[32,33,34]。 playing a vital role in the assembly of this complex [32][33][34]. The assembly process of the SMN复合物的组装过程是模块化的,涉及几种 complex is modular, involving the physical association of several SMN复合物蛋白质的物理结合。 complex proteins. SMN/Gemin8/Gemin7位于复合物的核心,募集其他蛋白质形成完整的组装体[34]。, situated at the core of the complex, recruit other proteins to form the complete assembly [34]. 作为分子伴侣,As a molecular companion, the SMN复合物促进剪接体小核糖核蛋白(snRNP)的组装和功能[35]。 complex facilitates the assembly and function of spliceosomal small ribonucleoprotein (snRNP) [35]. snRNP由富含 comprises U的小核RNA(snRNA)和7个小(Sm)或Sm样(LSm)蛋白组成,组装成细胞质中完整的茎环结构[34,36]。这种装配过程涉及多个-rich small nucleus RNA (snRNA) and 7 small (Sm) or Sm-like (LSm) proteins assembled into a complete stem ring structure in the cytoplasm [34][36]. This assembly process, which involves multiple SMN复杂组件,需要蜂窝位置偏移。 complex components, necessitates a cellular positional shift. 最初,Initially, snRNA(不包括U6)在细胞核中被RNA聚合酶II转录为前体RNA,在3'端包含一个额外的茎环结构,在7'端包含一个单甲基化的s (excluding U6) are transcribed in the nucleus by RNA polymerase II as precursor RNAs, containing an additional stem-loop structure at the 3′ end and a monomethylated m7GpppG(m5G)帽结构[37]。该前体 (m7G) cap structure at the 5′ end [37]. This precursor snRNA随后通过其5′帽结构与包含多种蛋白质的 subsequently binds to an snRNA输出复合物结合,导致其输出到细胞质中[38]。 export complex comprising multiple proteins through its 5′ cap structure, leading to its export into the cytoplasm [38]. 在细胞质中,In the cytoplasm, Sm和Sm样蛋白首先与氯化物电导调节蛋白(pICln)和蛋白精氨酸甲基转移酶5(PRMT5)复合物结合。然后将这些蛋白质预先排列到空间位置,并在加入SMN复合物后分离,SMN复合物将Sm和Sm样蛋白组装成snRNA上的蛋白质结合位点,形成具有七元环结构的完整snRNP[39,40]。 and Sm-like proteins first bind to the chloride conductance regulatory protein (pICln) and protein arginine methyltransferase 5 (PRMT5) complexes. These proteins are then prearranged into spatial positions and separated upon the addition of the SMN complex, which assembles the Sm and Sm-like proteins into protein binding sites on snRNAs, forming the complete snRNP with a seven-membered loop structure [39][40]. 最后,具有Sm核心的Finally, the snRNA在其5′末端经历甲基化,由三甲基鸟苷合酶1(TGS1)催化,形成m3G帽结构。该过程由SMN复合物和导入蛋白β介导,促进细胞核中剪接功能的表现[15,41]。这种组装过程很复杂,涉及不同的组件扮演独特的角色。本节的重点是简要概述 with the Sm core undergoes methylation at its 5′ end, catalyzed by trimethylguanosine synthase 1 (TGS1), to form an m3G cap structure. This process is mediated by the SMN complex and importin β, facilitating the performance of splicing functions in the nucleus [15][41]. This assembly process is complex and involves different components playing unique roles. The focus of this section is a brief overview of the role of DDX20 / Gemin3在此过程中的作用。 in this process. 首先,First and foremost, the SMN复合物在snRNP组装中起着至关重要的不可替代的作用,而DDX20 / complex serves a critical, nonreplaceable role in snRNP assembly, and DDX20/Gemin3对于复合物的稳定是必不可少的。在SMN复合物中,DDX20 / is indispensable for the stabilization of the complex. Within the SMN complex, DDX20/Gemin3与SMN蛋白以及 engages with SMN proteins, along with Gemin2,4和5结合。这种相互作用招募 4, and 5. This interaction recruits Gemin3作为SMN复合体的核心成分,促进其稳定性[42,43,44]。 as a core component of the SMN complex, promoting its stability [42][43][44]. A sumoylation modification of the core components of the SMN复合物的核心组分(包括 complex, including Gemin3)的sumo-, is necessary for this interaction. A desumoylation modification of Gemin3的脱硫修饰可能会减少与几种核心蛋白的结合,并可能影响细胞定位SMN复合物[29]。如图 could decrease binding to several core proteins and potentially impact the cellular localization SMN complex [29]. As shown in Figure 2所示,, Gemin3与其他几种蛋白质形成SMN复合物,并且各种蛋白质之间存在相互作用。 forms an SMN complex with several other proteins and there is interaction between the various proteins.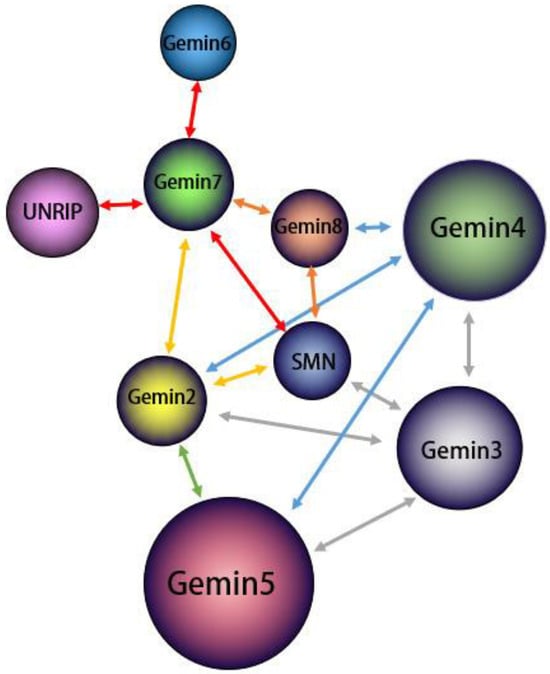
4. DDX20通过抑制转录因子抑制转录 Represses Transcription by Inhibiting Transcription Factors
近年来,In recent years, the role of DDX20在转录中的作用被报道得越来越多,并且已经报道了大量关于其作为转录调节剂作用的证据。DDX20主要通过与核受体类固醇生成因子-1(SF-1)相互作用来抑制转录[51]。 in transcription has been increasingly reported and considerable evidence regarding its role as a transcription regulator has been reported. DDX20 primarily suppresses transcription by interacting with the nuclear receptor steroidogenic factor-1 (SF-1) [51]. SF-1是转录因子核受体超家族的成员,是下丘脑-垂体-性腺轴和肾上腺皮质内分泌因子的关键调节因子[52]。, a member of the transcription factor nuclear receptor superfamily, is a crucial regulator of the hypothalamic–pituitary–gonadal axis and the endocrine factor of the adrenal cortex [52]. DDX20在产生类固醇的组织中高表达,类固醇组织也高表达 is highly expressed in steroid-producing tissues, which also highly express SF-1[10]。 [10]. DDX20通过其非保守的 directly interacts with the C端结构域直接与SF-1的C端抑制结构域相互作用,抑制SF-1的转录活性[20]。进一步的研究已经详细阐述了-terminal inhibitory domains of SF-1 via its nonconserved C-terminal domains to inhibit the transcriptional activity of SF-1 [20]. Further studies have elaborated on the specific mechanisms by which DDX20与 interacts with SF-1相互作用并抑制其转录活性的具体机制。转录因子SF-1的转录活性受其他转录因子、共调控因子和翻译后修饰的影响[53]。 and inhibits its transcriptional activity. The transcriptional activity of SF-1, a transcription factor, is affected by other transcription factors, coregulatory factors, and post-translational modifications [53]. Sumoylation, a ubiquitin-like modization是SF-1翻译后发生的泛素样修饰,可抑制SF-1激活靶基因表达的能力[54]。fication that occurs following SF-1 translation, inhibits the ability of SF-1 to activate target gene expression [54]. DDX20在与 enhances the sumoylation of SF-1蛋白直接相互作用以抑制其转录活性后,增强由活化的STATs蛋白抑制剂(一种 mediated by protein inhibitor of activated STATs proteins (a type of E3-SUMO连接酶)介导的SF-1的苏莫化[55]。这种相互作用也有助于 ligase) after direct interaction with the SF-1 protein to inhibit its transcriptional activity [55]. This interaction also facilitates the relocalization of SF-1重新定位到离散的核小体或病变。探索这种抑制机制表明,组蛋白去乙酰化酶参与转录因子 to discrete nucleosomes or lesions. Exploring this inhibitory mechanism revealed that Histone deacetylase is involved in sumoylation modification of transcription factor SF-1的sumo-ization修饰,并且与核心加压因子的相互作用较少;然而,它可能在苏莫酰化过程中作为E3连接酶起作用[56]。 and interacts less with the corepressor; however, it may function as an E3 ligase during sumoylation [56]. SF-1在节肢动物中具有同源基因,称为 has a homologous gene in arthropods, known as FTZ-F1( (fushi tarazu因子-1)[57]。一项研究证实, factor-1) [57]. A study confirmed that DDX20通过酵母双杂交测定法与副苜蓿中的 interacts with FTZ-F1相互作用,并报道DDX20可以通过类似于RNAi抑制SF-1的机制抑制 in paralfalfa via a yeast two-hybrid assay and reported that DDX20 can suppress FTZ-F1 expression via a mechanism similar to that of SF-1 inhibition by RNAi [58]. FTZ-F1表达[58]。 is closely related to vitellogenin (VTG) and is involved in the development of vitelline in the ovary and can influence the secretion of several endocrine hormones [59]. The inhibition of FTZ-F1与卵黄原素(VTG)密切相关,参与卵巢中卵黄素的发育,可影响几种内分泌激素的分泌[59]。通过 via DDX1抑制FTZ-F20最终会影响卵巢发育。 eventually affects ovarian development. 叉头转录因子(The Forkhead transcription factor (FOXL)2是与) 2, a transcription factor closely related to FTZ-F1和DDX20密切相关的转录因子,在调节卵巢发育方面也起着重要作用。在卵巢中,FOXL2参与调节胆固醇和类固醇代谢、细胞凋亡、活性氧解毒和细胞增殖[60]。最初,发现 and DDX20, also plays a significant role in regulating ovarian development. In the ovary, FOXL2 is involved in regulating cholesterol and steroid metabolism, apoptosis, reactive oxygen species detoxification, and cell proliferation [60]. Initially, it was discovered that DDX20与 interacted with FOXL2相互作用,它们在细胞中的共表达增强了FOXL2介导的卵巢细胞死亡[61]。随后的研究证实, and their coexpression in cells enhanced FOXL2-mediated ovarian cell death [61]. Subsequent studies confirmed that FOXL2与 interacts with DDX20和 and FTZ-F1相互作用,不仅通过DDX20调节滤泡细胞凋亡来下调VTG合成,而且还可能通过 and not only downregulates VTG synthesis by regulating follicular cell apoptosis through DDX20 but also potentially regulates the steroidogenic pathway through FTZ-F1调节类固醇生成途径。. 除In addition to F-1外,已发现DDX20与另外两种转录因子相互作用,与SF-1相比,通过不同的机制抑制其活性。首先,DDX20可以通过其C端结构域与有丝分裂的Ets转录抑制剂(PE-1 / METS)结合。Ets是一种转录因子,可作为核靶点激活Ras-MAPK信号通路,而Ets家族的另一个成员, DDX20 has been discovered to interact with two other transcription factors to inhibit their activity by different mechanisms compared with that used with SF-1. First, DDX20 can engage with the mitotic Ets transcription inhibitor (PE-1/METS) via its C-terminal domain. Ets is a transcription factor that serves as a nuclear target to activate the Ras–MAPK signaling pathway, while PE-1/METS则作为Ets抑制剂抑制Ras依赖性Ets靶基因增殖,从而导致巨噬细胞生长停滞[62]。, another member of the Ets family, acts as an Ets inhibitor to repress the Ras-dependent proliferation of Ets target genes, thus causing macrophage growth arrest [62]. DDX20通过其C端结构域与 interacts with the N-terminal domain of PE-1/METS的N端结构域相互作用,具有抑制作用。在此过程中,DDX20还募集组蛋白去乙酰化酶 via its C-terminal domain, which has an inhibitory effect. In this process, DDX20 also recruits factors such as histone deacetylase HDAC-2和HDAC5等因子[63]。然而,有趣的是,这种由 and HDAC5 [63]. However, interestingly, this transcriptional inhibition mediated by DDX20介导的转录抑制仅针对单个启动子,例如 targets only individual promoters, such as c-myc和cdc2,而不影响Ets三元复合物促进的转录[64]。 and cdc2, without affecting Ets ternary complex-facilitated transcription [64]. Thus, DDX20 does not undergo sumoacylation as DDX5 and its transcription regulation activity do not solely rely on a single intrinsic function but involve multiple mechanisms, many of which depend on its unique noncore C-terminal domain. This multifaceted approach to transcriptional regulation reinforces the complexity of the function of DDX20 in this essential cellular process.5. Biogenesis of DDX20 and miRNA
DDX20/Gemin3 also contributes to the maturation of miRNA. The biogenesis of miRNA begins with the transcription of miRNA genes into primary miRNA (pri-miRNA) by RNA polymerase II. The pri-miRNA is subsequently processed into a precursor miRNA (pre-miRNA) of ~70 nucleotides by a complex containing the RNase III endonuclease Drosha and a protein containing a double-stranded RNA-binding structural domain (dsRBD) named Pasha (DGCR8) [77,78][65][66]. Subsequently, the stem–loop-structured pre-miRNAs are transported into the cytoplasm, which is dependent on the GTP-dependent transport protein Exportin-5 (Exp5) [79][67]. In the cytoplasm, the RNase III endonuclease Dicer processes pre-miRNA into an siRNA–miRNA duplex of ~22 nucleotides. The mature miRNA strand is subsequently retained in the RNA-induced silencing complex (RISC) along with other proteins [80][68]. Although most Gemin3 and Gemin4 are components of the SMN complex, the complexes of Gemin3 and Gemin4 isolated from HeLa and neuronal cells are not part of the SMN complex. Instead, these complexes coprecipitate with polyribosomes [71,81,82][69][70][71]. Numerous studies have established the connection of Gemin3/DP103 and Gemin4 with the Argonaute (Ago) protein family member Argonaute [60,83,84][60][72][73]. Figure 4 shows the mIRNAde1 maturation process and the role Gemin3 plays in it.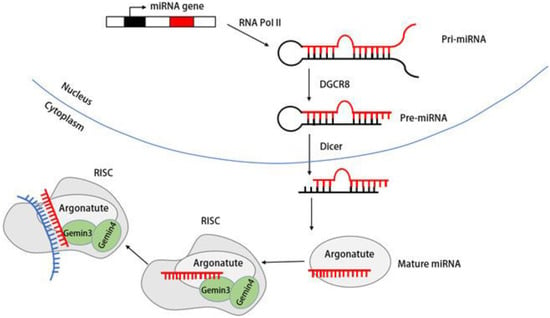
6. Functions of DDX20 in the Innate Immune Signaling Pathway
6.1. Effect of DDX20 on the NF-κB Signaling Pathway
The NF-κB family comprises six distinct components. When activated, they produce various proinflammatory factors that regulate inflammation and significantly contribute to innate immunity [87,88][77][78]. The NF-κB signaling pathway controls the expression of proinflammatory cytokines and anti-infective factors, including TNF-α, interleukin-1 (IL-1), IL-6, IL-8, adhesion molecules, and cc chemokine ligand 5 (CCL5) [89][79]. In addition, NF-κB signaling pathway governs cellular processes such as cell proliferation, differentiation, and apoptosis [90,91][80][81]. While the NF-κB signaling pathway has an antiapoptotic role, its dysregulation has been implicated in the pathogenesis of most human malignancies [92][82]. Therefore, manipulating the NF-κB signaling pathway can provide a path for developing novel methods to combat diseases such as cancer [93][83]. For instance, alterations in DDX20 expression in cancer tissues can affect NF-κB activity, leading to cancer development [94][84]. DDX20 can have two distinct effects on the NF-κB signaling pathway. Numerous studies have reported that DDX20 does not directly affect NF-κB activity but rather acts through a naturally occurring small non-coding RNA (miRNA) intermediate. The present study demonstrated that DDX20 can modulate the signaling of the NF-κB pathway through miRNA-22, miRNA-140-3p, and miR-222 [8,95][8][85]. Additionally, miR-361-5p was found to regulate DDX20, thereby influencing NF-κB pathway signaling [13]. First, DDX20 can inhibit NF-κB activity. DDX20 and miRNAs together form a ribonucleoprotein complex, and miRNA library screening has revealed that several miRNAs can inhibit NF-κB activation. This inhibition prevents the activation of the NF-κB signaling pathway [8]. It has been well established that miRNAs can inhibit NF-κB activity by regulating the expression of two NF-κB coactivators, namely nuclear receptor coactivator protein 1 (NCOA1) and nuclear receptor-interacting protein 1 (NRIP1) [96][86]. Conversely, DDX20 can also enhance NF-κB activity, notably by impairing miRNA function by decreasing its own expression, leading to impaired NF-κB inhibitory miRNA function [8,97][8][87]. Another way DDX20 can inhibit the NF-κB signaling pathway is through miRNA-140 dysfunction, which increases expression of its downstream target gene Dnmt1 and the methylation of CpG islands in the promoter region of metallothionein (MTs) and decreases MT expression, leading to enhanced NF-κB activity [9]. Second, DDX20 can boost the NF-κB signaling pathway by enhancing the phosphorylation of TAK 1, where it acts as a cofactor of TAK1, thus enhancing the activity of the NF-κB signaling pathway [98][88]. More details regarding this aspect will be discussed in the subsequent sections.6.2. DDX20 Affects p53 Signaling Pathway Conduction
The TP53 gene encodes the key p53 transcription factor and evolutionarily conserved tumor suppressor involved in maintaining genomic stability [99][89]. To accomplish this, it activates DNA repair responses and initiates apoptosis in damaged host cells [100][90]. Its activation controls core programs such as cell cycle arrest and apoptosis [101][91]. In addition, p53 is closely related to immune responses as well as various inflammatory diseases [102][92]. In fact, the effect of DDX20 on the p53 signaling pathway can be realized by directly affecting the pivotal p53 protein. Changes in DDX20 expression can consequently impact the organism’s state through the p53 signaling pathway. Reportedly, DDX20 interacts with p53 protein through its C-terminal structural domain [1]. The normal expression of DDX20 stabilizes motor neurons and uses genomic stability and regulates Mdm2 splicing to restrain the p53 signaling pathway, thereby preserving normal neural development [4]. In this context, several cytokines, such as oligodendrocyte transcription factor 2 (Olig2) and Epstein–Barr (EB) nuclear antigen 3C (EBNA3C), can directly interact with DDX20 to stabilize its expression. This interaction inhibits transcription and apoptosis caused by the p53 signaling pathway and its downstream genes within the organism [4,67][4][93].7. DDX20 Plays Different Roles in Cancers through the NF-κB Signaling Pathway
DDX20 not only plays a role in the invasion of multiple pathogens but also plays an active role in the case of multiple tumorigenesis. In breast cancer, DDX20 is involved in cell signaling pathway activity, which is a key factor in tumorigenesis. The NF-κB signaling pathway is more closely linked to tumor development, and, reportedly, it can improve cancer cell survival, promote cancer cell angiogenesis and migration, and has other characteristics alongside being associated with the poor prognosis of cancer diseases [106][94]. It is because of the role of the NF-κB signaling pathway in tumor growth that it can be used to inhibit carcinogenesis [107][95]. DDX20 is an important cofactor for the phosphorylation of transforming growth factor-β-activated kinase-1 (TAK1) by NF-κB-activated IκB kinase 2 (IKK2), enhancing the activity of the NF-κB signaling pathway by stimulating TAK1 phosphorylation [108][96]. TAK1 is a member of the mitogen-activated protein kinase (MAPK) family that plays a key regulatory role as an upstream component of the NF-κB signaling pathway [109][97]. Enhanced NF-κB signaling pathway activation increases in the expression of two downstream products, matrix metalloproteinase 9 (MMP9) and multidrug resistance gene 1 (MDR1), and, as the chief role of MMP9 is to degrade the extracellular matrix, this change further increases tumor metastasis and drug resistance [19,110][19][98]. Conversely, enhanced NF-κB activity and increased downstream MMP9 expression would also lead to increased DDX20 expression. Thus, the establishment of the DDX20–NF-κB–MMP9 axis could better reveal the mechanism by which DDX20 can promote cancer development [69][99]. In addition, DDX20 may exhibit an miRNA-processing role in breast cancer. A group of studies reported that DDX20 exhibited a negative correlation with an miRNA, namely miR-222, suggesting that DDX20 affects the NF-κB activity through miR-222 to promote breast cancer development [95][85]. Based on this property of DDX20 in breast cancer, researchers believe that DDX20 can serve as an active alternative to certain anticancer drugs. DDX20 enhances the sumolylation modification of YAP, thereby increasing YP-TEAD dependence and statin sensitivity in patients with triple-negative breast cancer [16]. Statins, such as simvastatin (SMV), are cholesterol-reducing lipophilic statins that inhibit DDX20 expression by inhibiting 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), which is positively associated with DDX20 [103][100]. Figure 5A demonstrates that DDX20 affects tumorigenesis and development through the NF-κB signaling pathway. Further studies have reported that simvastatin downregulates DDX20 not only through the classical mevaleric acid pathway and the downstream component of RHoA but also through the miRNA-mediated nonclassical pathway [111,112][101][102]. Simvastatin ultimately inhibits breast cancer by decreasing DDX20 expression. In addition to its role through the NF-κB signaling pathway and miR-222, DDX20 acts through the cellular redox pathway and Wingless/Integrated(Wnt) signaling pathway. In cancer stem cells (CSCS), DDX20 drives a positive feedback loop wherein DDX20 promotes its transcriptional regulation via transcription factor 4 (TCF 4) to stimulates the aggressiveness of breast cancer via Wnt/beta-catenin signaling [113][103]. Another example of the carcinogenic effect of DDX20 is in prostate cancer. High DDX20 expression in the tumor tissue of patients with prostate cancer also enhances tumor growth and metastasis via the DDX20–NF-κB–MMP9 axis [11]. These results suggest a role of DDX20 in tumor metastasis.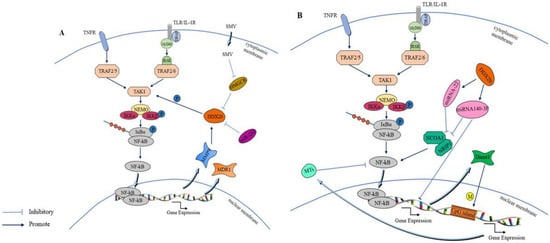
8. Functions in Viral Infection
Early research identified the capacity of DDX20 to interact with two vitally encoded nuclear antigens of the EB virus (EBV), EBNA2 and EBNA3C, and modulate the transcription of viral and cellular genes [68][110]. EBV is a lymphocryptovirus (LCV) herpesvirus that predominantly infects B lymphocytes and is noted for its ability to maintain long-term latent infections in the body while expressing a limited number of “latent” genes [119,120][111][112]. According to the report, EBNA2 can serve as a transcriptional activator of transformative viral and cellular genes by regulating two EBNA2-regulated viral promoters (TP1 and LMP/TP2 promoter) following its binding to the homologous promoter element RBPJkappa [121,122][113][114]. This promoter activation by EBNA2 is facilitated by cellular enhancer binding proteins and EBV nucleoproteins [123,124][115][116]. Later studies revealed that DDX20 can bind EBNA2 and survival motor neuron (SMN) proteins via its C-terminal structural domain, reporting that EBNA2 targets the spliceosome complex to release the SMN protein after binding DDX20, which ultimately acts as a coactivator in RNA polymerase II transcriptional complexes on the LMP1 promoter [125][117]. An additional finding suggests that, although RBPJkappa is necessary for EBNA2 transactivation, it is insufficient and it needs to be achieved through the EBNA2 and SMN proteins, underscoring the crucial role of DDX20 [125][117]. Conversely, EB nuclear antigen 3C (EBNA3C) is an important latent antigen that induces B lymphocyte immortalization in EBV through its contribution to viral pathogenicity. This is achieved via the putative bZIP structural domain at the N terminus of the protein and its interaction with cellular transcription factors RBPJkappa and HDAC1 to regulate transcriptional activation [126,127][118][119]. Additionally, EBNA3C facilitates the transcriptional reprogramming of various host cell genes, which is associated with the lengthy latency of EBV [128][120]. Similarly, DDX20 interacts with EBNA3C via its C-terminal structural domain. DDX20 can be stabilized by EBNA3C to form a complex with p53, which subsequently blocks p53-mediated transcriptional activity and apoptosis [67][93]. Moreover, based on related studies, EBNA2 and EBNA3C can reportedly disrupt the interaction between DDX20 and the transcription factor METS, thereby activating cellular proto-oncogenes [129][121]. This connection between DDX20 and EBV led to the initial discovery of a link between DDX20 and cancer. In addition to EBV, DDX20 has significant connections with other viruses. DDX20 expression has previously been identified to vary in tumor tissues. In terms of changes in its expression upon viral infection, researchers reported that DDX20 is differentially expressed at distinct stages of human immunodeficiency virus type 1 (HIV-1) infection and found that DDX20 interacts with the HIV-1 coprotein Vpr as early as 2012 [76][122]. Further analysis of global miRNA and mRNA expression through microarray and quantitative reverse transcription polymerase chain reaction in well-characterized HIV-1 latently and actively infected cells revealed that DDX20 was significantly upregulated in the former [130][123]. Conversely, a downregulation of DDX20 expression was detected during HIV-1 replication. DDX20 was directly degraded by the vRNA translation product of HIV-1, the Vpr protein, via the DCAF1/DDB1/CUL4 E3 ubiquitin ligase-mediated degradation pathway [131][124]. Owing to these alterations in DDX20 expression, it is thought to play a part in HIV-1 replication. However, the precise mechanism underlying the potential inhibitory effect of DDX20 remains unclear, thus necessitating further research. DDX20 can enhance Interferon regulatory factor 3 (IRF3) phosphorylation levels by promoting the interaction between TBK1 and IRF3, which further promotes IFN-β expression, ultimately inhibiting the replication of VSV and HSV-1 through INF-stimulated genes (ISG) [2]. This opens up avenues for investigating the role of DDX20 in innate immunity research. Thus, although the role of DDX20 in suppressing viral infection and innate immunity has not been extensively studied, it has great research potential.References
- Curmi, F.; Cauchi, R.J. The multiple lives of DEAD-box RNA helicase DP103/DDX20/Gemin3. Biochem. Soc. Trans. 2018, 46, 329–341.
- Wang, X. The Role and Mechanisms of DEAD-Box RNA Helicase DDX20 on Antiviral Innate Immunity. Master’s Thesis, Soochow University, Suzhou, China, 2019.
- Mouillet, J.-F.; Yan, X.; Ou, Q.; Jin, L.; Muglia, L.J.; Crawford, P.A.; Sadovsky, Y. DEAD-box protein-103 (DP103, Ddx20) is essential for early embryonic development and modulates ovarian morphology and function. Endocrinology 2008, 149, 2168–2175.
- Bizen, N.; Bepari, A.K.; Zhou, L.; Abe, M.; Sakimura, K.; Ono, K.; Takebayashi, H. Ddx20, an Olig2 binding factor, governs the survival of neural and oligodendrocyte progenitor cells via proper Mdm2 splicing and p53 suppression. Cell Death Differ. 2022, 29, 1028–1041.
- Simankova, A.; Bizen, N.; Saitoh, S.; Shibata, S.; Ohno, N.; Abe, M.; Sakimura, K.; Takebayashi, H. Ddx20, DEAD box helicase 20, is essential for the differentiation of oligodendrocyte and maintenance of myelin gene expression. Glia 2021, 69, 2559–2574.
- Hobani, Y.H.; Almars, A.I.; Alelwani, W.; Toraih, E.A.; Nemr, N.A.; Shaalan, A.A.M.; Fawzy, M.S.; Attallah, S.M. Genetic Variation in DEAD-Box Helicase 20 as a Putative Marker of Recurrence in Propensity-Matched Colon Cancer Patients. Genes 2022, 13, 1404.
- Minasaki, R.; Puoti, A.; Streit, A. The DEAD-box protein MEL-46 is required in the germ line of the nematode Caenorhabditis elegans. BMC Dev. Biol. 2009, 9, 35.
- Takata, A.; Otsuka, M.; Kojima, K.; Yoshikawa, T.; Kishikawa, T.; Ijichi, H.; Hirata, Y.; Tateishi, K.; Yoshida, H.; Omata, M.; et al. Abstract 3978: DDX20, a suppressor of hepatocarcinogenesis, controls NF-κB activity through regulating the function of miRNA-22 and miRNA-140-3p targeting transcriptional coactivators. Cancer Res. 2011, 71, 3978.
- Takata, A.; Otsuka, M.; Yoshikawa, T.; Kishikawa, T.; Hikiba, Y.; Obi, S.; Goto, T.; Kang, Y.J.; Maeda, S.; Yoshida, H.; et al. MicroRNA-140 acts as a liver tumor suppressor by controlling NF-κB activity by directly targeting DNA methyltransferase 1 (Dnmt1) expression. Hepatology 2013, 57, 162–170.
- Ou, Q.; Mouillet, J.F.; Yan, X.; Dorn, C.; Crawford, P.A.; Sadovsky, Y. The DEAD box protein DP103 is a regulator of steroidogenic factor-1. Mol. Endocrinol. 2001, 15, 69–79.
- Chen, W.; Zhou, P.; Li, X. High expression of DDX20 enhances the proliferation and metastatic potential of prostate cancer cells through the NF-κB pathway. Int. J. Mol. Med. 2016, 37, 1551–1557.
- Yang, Y.; Yang, M.; Pang, H.; Qiu, Y.; Sun, T.; Wang, T.; Shen, S.; Wang, W. A Macrophage Differentiation-Mediated Gene: DDX20 as a Molecular Biomarker Encompassing the Tumor Microenvironment, Disease Staging, and Prognoses in Hepatocellular Carcinoma. Oxidative Med. Cell. Longev. 2022, 2022, 9971776.
- Wang, Q.; Ye, Y.; Lin, R.; Weng, S.; Cai, F.; Zou, M.; Niu, H.; Ge, L.; Lin, Y. Analysis of the expression, function, prognosis and co-expression genes of DDX20 in gastric cancer. Comput. Struct. Biotechnol. J. 2020, 18, 2453–2462.
- Vychytilova-Faltejskova, P.; Svobodova Kovarikova, A.; Grolich, T.; Prochazka, V.; Slaba, K.; Machackova, T.; Halamkova, J.; Svoboda, M.; Kala, Z.; Kiss, I.; et al. MicroRNA Biogenesis Pathway Genes Are Deregulated in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 4460.
- Kroiss, M.; Schultz, J.; Wiesner, J.; Chari, A.; Sickmann, A.; Fischer, U. Evolution of an RNP assembly system: A minimal SMN complex facilitates formation of UsnRNPs in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2008, 105, 10045–10050.
- Lai, X.; Wang, T.; Tan, T.Z.; Casey, P.J.; Hong, W.; Sudol, M.; Tergaonkar, V.; Lee, S.C.; Kumar, A.P. Abstract A28: DEAD-box RNA helicase DP103 enhances YAP sumoylation for YAP-TEAD dependence and statin sensitivity in triple-negative breast cancer. Mol. Cancer Res. 2020, 18, A28.
- Ali, M.A.M. DEAD-box RNA helicases: The driving forces behind RNA metabolism at the crossroad of viral replication and antiviral innate immunity. Virus Res. 2021, 296, 198352.
- Schütz, P.; Karlberg, T.; van den Berg, S.; Collins, R.; Lehtiö, L.; Högbom, M.; Holmberg-Schiavone, L.; Tempel, W.; Park, H.W.; Hammarström, M.; et al. Comparative structural analysis of human DEAD-box RNA helicases. PLoS ONE 2010, 5, e12791.
- Ali, M.A.M. The DEAD-box protein family of RNA helicases: Sentinels for a myriad of cellular functions with emerging roles in tumorigenesis. Int. J. Clin. Oncol. 2021, 26, 795–825.
- Yan, X.; Mouillet, J.F.; Ou, Q.; Sadovsky, Y. A novel domain within the DEAD-box protein DP103 is essential for transcriptional repression and helicase activity. Mol. Cell Biol. 2003, 23, 414–423.
- Coady, T.H.; Lorson, C.L. SMN in spinal muscular atrophy and snRNP biogenesis. Wiley Interdiscip. Rev. RNA 2011, 2, 546–564.
- Burlet, P.; Huber, C.; Bertrandy, S.; Ludosky, M.A.; Zwaenepoel, I.; Clermont, O.; Roume, J.; Delezoide, A.L.; Cartaud, J.; Munnich, A.; et al. The distribution of SMN protein complex in human fetal tissues and its alteration in spinal muscular atrophy. Hum. Mol. Genet. 1998, 7, 1927–1933.
- Charroux, B.; Pellizzoni, L.; Perkinson, R.A.; Shevchenko, A.; Mann, M.; Dreyfuss, G. Gemin3: A novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell. Biol. 1999, 147, 1181–1194.
- Pellizzoni, L.; Yong, J.; Dreyfuss, G. Essential role for the SMN complex in the specificity of snRNP assembly. Science 2002, 298, 1775–1779.
- Lemm, I.; Girard, C.; Kuhn, A.N.; Watkins, N.J.; Schneider, M.; Bordonné, R.; Lührmann, R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol. Biol. Cell 2006, 17, 3221–3231.
- Trinkle-Mulcahy, L.; Boulon, S.; Lam, Y.W.; Urcia, R.; Boisvert, F.M.; Vandermoere, F.; Morrice, N.A.; Swift, S.; Rothbauer, U.; Leonhardt, H.; et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 2008, 183, 223–239.
- Stanek, D.; Neugebauer, K.M. The Cajal body: A meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma 2006, 115, 343–354.
- Husedzinovic, A.; Oppermann, F.; Draeger-Meurer, S.; Chari, A.; Fischer, U.; Daub, H.; Gruss, O.J. Phosphoregulation of the human SMN complex. Eur. J. Cell Biol. 2014, 93, 106–117.
- Riboldi, G.M.; Faravelli, I.; Kuwajima, T.; Delestrée, N.; Dermentzaki, G.; De Planell-Saguer, M.; Rinchetti, P.; Hao, L.T.; Beattie, C.C.; Corti, S.; et al. Sumoylation regulates the assembly and activity of the SMN complex. Nat. Commun. 2021, 12, 5040.
- Lafarga, V.; Tapia, O.; Sharma, S.; Bengoechea, R.; Stoecklin, G.; Lafarga, M.; Berciano, M.T. CBP-mediated SMN acetylation modulates Cajal body biogenesis and the cytoplasmic targeting of SMN. Cell. Mol. Life Sci. CMLS 2018, 75, 527–546.
- Meier, I.D.; Walker, M.P.; Matera, A.G. Gemin4 is an essential gene in mice, and its overexpression in human cells causes relocalization of the SMN complex to the nucleoplasm. Biol. Open 2018, 7, bio032409.
- Fallini, C.; Bassell, G.J.; Rossoll, W. Spinal muscular atrophy: The role of SMN in axonal mRNA regulation. Brain Res. 2012, 1462, 81–92.
- Patterson, W.L., 3rd; Georgel, P.T. Breaking the cycle: The role of omega-3 polyunsaturated fatty acids in inflammation-driven cancers. Biochem. Cell Biol. Biochim. Biol. Cell. 2014, 92, 321–328.
- Meister, G.; Bühler, D.; Pillai, R.; Lottspeich, F.; Fischer, U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 2001, 3, 945–949.
- Battle, D.J.; Kasim, M.; Wang, J.; Dreyfuss, G. SMN-independent subunits of the SMN complex. Identification of a small nuclear ribonucleoprotein assembly intermediate. J. Biol. Chem. 2007, 282, 27953–27959.
- Will, C.L.; Lührmann, R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001, 13, 290–301.
- Massenet, S.; Pellizzoni, L.; Paushkin, S.; Mattaj, I.W.; Dreyfuss, G. The SMN complex is associated with snRNPs throughout their cytoplasmic assembly pathway. Mol. Cell. Biol. 2002, 22, 6533–6541.
- Matera, A.G.; Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014, 15, 108–121.
- Martinez-Salas, E.; Embarc-Buh, A.; Francisco-Velilla, R. Emerging Roles of Gemin5: From snRNPs Assembly to Translation Control. Int. J. Mol. Sci. 2020, 21, 3868.
- Li, D.K.; Tisdale, S.; Lotti, F.; Pellizzoni, L. SMN control of RNP assembly: From post-transcriptional gene regulation to motor neuron disease. Semin. Cell Dev. Biol. 2014, 32, 22–29.
- Mouaikel, J.; Narayanan, U.; Verheggen, C.; Matera, A.G.; Bertrand, E.; Tazi, J.; Bordonné, R. Interaction between the small-nuclear-RNA cap hypermethylase and the spinal muscular atrophy protein, survival of motor neuron. EMBO Rep. 2003, 4, 616–622.
- Otter, S.; Grimmler, M.; Neuenkirchen, N.; Chari, A.; Sickmann, A.; Fischer, U. A Comprehensive Interaction Map of the Human Survival of Motor Neuron (SMN) Complex. J. Biol. Chem. 2007, 282, 5825–5833.
- Charroux, B.; Pellizzoni, L.; Perkinson, R.A.; Yong, J.; Shevchenko, A.; Mann, M.; Dreyfuss, G. Gemin4. A novel component of the SMN complex that is found in both gems and nucleoli. J. Cell Biol. 2000, 148, 1177–1186.
- Cauchi, R.J. SMN and Gemins: ‘we are family’ … or are we?: Insights into the partnership between Gemins and the spinal muscular atrophy disease protein SMN. BioEssays News Rev. Mol. Cell. Dev. Biol. 2010, 32, 1077–1089.
- Feng, W.; Gubitz, A.K.; Wan, L.; Battle, D.J.; Dostie, J.; Golembe, T.J.; Dreyfuss, G. Gemins modulate the expression and activity of the SMN complex. Hum. Mol. Genet. 2005, 14, 1605–1611.
- Cauchi, R.J.; Davies, K.E.; Liu, J.L. A motor function for the DEAD-box RNA helicase, Gemin3, in Drosophila. PLoS Genet. 2008, 4, e1000265.
- Almstead, L.L.; Sarnow, P. Inhibition of U snRNP assembly by a virus-encoded proteinase. Genes. Dev. 2007, 21, 1086–1097.
- Shpargel, K.B.; Matera, A.G. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc. Natl. Acad. Sci. USA 2005, 102, 17372–17377.
- Borg, R.M.; Fenech Salerno, B.; Vassallo, N.; Bordonne, R.; Cauchi, R.J. Disruption of snRNP biogenesis factors Tgs1 and pICln induces phenotypes that mirror aspects of SMN-Gemins complex perturbation in Drosophila, providing new insights into spinal muscular atrophy. Neurobiol. Dis. 2016, 94, 245–258.
- Cacciottolo, R.; Ciantar, J.; Lanfranco, M.; Borg, R.M.; Vassallo, N.; Bordonné, R.; Cauchi, R.J. SMN complex member Gemin3 self-interacts and has a functional relationship with ALS-linked proteins TDP-43, FUS and Sod1. Sci. Rep. 2019, 9, 18666.
- Fuller-Pace, F.V. DExD/H box RNA helicases: Multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006, 34, 4206–4215.
- Parker, K.L.; Schimmer, B.P. Steroidogenic factor 1: A key determinant of endocrine development and function. Endocr. Rev. 1997, 18, 361–377.
- Zhao, L.; Kim, K.W.; Ikeda, Y.; Anderson, K.K.; Beck, L.; Chase, S.; Tobet, S.A.; Parker, K.L. Central nervous system-specific knockout of steroidogenic factor 1 results in increased anxiety-like behavior. Mol. Endocrinol. 2008, 22, 1403–1415.
- Lee, J.; Yang, D.J.; Lee, S.; Hammer, G.D.; Kim, K.W.; Elmquist, J.K. Nutritional conditions regulate transcriptional activity of SF-1 by controlling sumoylation and ubiquitination. Sci. Rep. 2016, 6, 19143.
- Lee, M.B.; Lebedeva, L.A.; Suzawa, M.; Wadekar, S.A.; Desclozeaux, M.; Ingraham, H.A. The DEAD-box protein DP103 (Ddx20 or Gemin-3) represses orphan nuclear receptor activity via SUMO modification. Mol. Cell. Biol. 2005, 25, 1879–1890.
- Treuter, E.; Venteclef, N. Transcriptional control of metabolic and inflammatory pathways by nuclear receptor SUMOylation. Biochim. Biophys. Acta 2011, 1812, 909–918.
- Hoivik, E.A.; Lewis, A.E.; Aumo, L.; Bakke, M. Molecular aspects of steroidogenic factor 1 (SF-1). Mol. Cell. Endocrinol. 2010, 315, 27–39.
- Yao, C.; Sun, Y.; Zhang, Z.; Jia, X.; Zou, P.; Wang, Y. Integration of RNAi and RNA-seq uncovers the regulation mechanism of DDX20 on vitellogenin expression in Scylla paramamosain. Comp. Biochem. Physiol. Part D Genom. Proteom. 2022, 44, 101028.
- Li, Q.; Xie, J.; He, L.; Wang, Y.; Yang, H.; Duan, Z.; Wang, Q. FOXL2 down-regulates vitellogenin expression at mature stage in Eriocheir sinensis. Biosci. Rep. 2015, 35, e00278.
- Pisarska, M.D.; Bae, J.; Klein, C.; Hsueh, A.J. Forkhead l2 is expressed in the ovary and represses the promoter activity of the steroidogenic acute regulatory gene. Endocrinology 2004, 145, 3424–3433.
- Lee, K.; Pisarska, M.D.; Ko, J.J.; Kang, Y.; Yoon, S.; Ryou, S.M.; Cha, K.Y.; Bae, J. Transcriptional factor FOXL2 interacts with DP103 and induces apoptosis. Biochem. Biophys. Res. Commun. 2005, 336, 876–881.
- Sawka-Verhelle, D.; Escoubet-Lozach, L.; Fong, A.L.; Hester, K.D.; Herzig, S.; Lebrun, P.; Glass, C.K. PE-1/METS, an antiproliferative Ets repressor factor, is induced by CREB-1/CREM-1 during macrophage differentiation. J. Biol. Chem. 2004, 279, 17772–17784.
- Klappacher, G.W.; Lunyak, V.V.; Sykes, D.B.; Sawka-Verhelle, D.; Sage, J.; Brard, G.; Ngo, S.D.; Gangadharan, D.; Jacks, T.; Kamps, M.P.; et al. An induced Ets repressor complex regulates growth arrest during terminal macrophage differentiation. Cell 2002, 109, 169–180.
- Hester, K.D.; Verhelle, D.; Escoubet-Lozach, L.; Luna, R.; Rose, D.W.; Glass, C.K. Differential repression of c-myc and cdc2 gene expression by ERF and PE-1/METS. Cell Cycle 2007, 6, 1594–1604.
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297.
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240.
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98.
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744.
- Mourelatos, Z.; Dostie, J.; Paushkin, S.; Sharma, A.; Charroux, B.; Abel, L.; Rappsilber, J.; Mann, M.; Dreyfuss, G. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes. Dev. 2002, 16, 720–728.
- Dostie, J.; Mourelatos, Z.; Yang, M.; Sharma, A.; Dreyfuss, G. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA 2003, 9, 180–186.
- Nelson, P.T.; Hatzigeorgiou, A.G.; Mourelatos, Z. miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA 2004, 10, 387–394.
- Hutvágner, G.; Zamore, P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science 2002, 297, 2056–2060.
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197.
- Pillai, R.S.; Bhattacharyya, S.N.; Filipowicz, W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007, 17, 118–126.
- Behm-Ansmant, I.; Rehwinkel, J.; Doerks, T.; Stark, A.; Bork, P.; Izaurralde, E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes. Dev. 2006, 20, 1885–1898.
- Murashov, A.K.; Chintalgattu, V.; Islamov, R.R.; Lever, T.E.; Pak, E.S.; Sierpinski, P.L.; Katwa, L.C.; Van Scott, M.R. RNAi pathway is functional in peripheral nerve axons. FASEB J. 2007, 21, 656–670.
- Bollrath, J.; Greten, F.R. IKK/NF-kappaB and STAT3 pathways: Central signalling hubs in inflammation-mediated tumour promotion and metastasis. EMBO Rep. 2009, 10, 1314–1319.
- Sun, S.C. The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017, 17, 545–558.
- Gilmore, T.D. Introduction to NF-kappaB: Players, pathways, perspectives. Oncogene 2006, 25, 6680–6684.
- Guttridge, D.C.; Albanese, C.; Reuther, J.Y.; Pestell, R.G.; Baldwin, A.S., Jr. NF-kappaB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol. Cell. Biol. 1999, 19, 5785–5799.
- Patel, M.; Horgan, P.G.; McMillan, D.C.; Edwards, J. NF-κB pathways in the development and progression of colorectal cancer. Transl. Res. J. Lab. Clin. Med. 2018, 197, 43–56.
- Verzella, D.; Pescatore, A.; Capece, D.; Vecchiotti, D.; Ursini, M.V.; Franzoso, G.; Alesse, E.; Zazzeroni, F. Life, death, and autophagy in cancer: NF-κB turns up everywhere. Cell Death Dis. 2020, 11, 210.
- Motolani, A.; Martin, M.; Sun, M.; Lu, T. 6.19—NF-κB and Cancer Therapy Drugs. In Comprehensive Pharmacology; Kenakin, T., Ed.; Elsevier: Oxford, UK, 2022; pp. 351–363.
- Tao, Y.; Zhou, J.; Wang, Z.; Tao, H.; Bai, J.; Ge, G.; Li, W.; Zhang, W.; Hao, Y.; Yang, X.; et al. Human bone mesenchymal stem cells-derived exosomal miRNA-361-5p alleviates osteoarthritis by downregulating DDX20 and inactivating the NF-κB signaling pathway. Bioorganic Chem. 2021, 113, 104978.
- Cai, W.; Wang, C.; Goh, J.N.; Loo, S.Y.; Yap, C.T.; Kumar, A.P. DEAD-box RNA Helicases: The microRNA managers of breast cancer. RNA Dis. 2015, 2, e846.
- Takata, A.; Otsuka, M.; Kojima, K.; Yoshikawa, T.; Kishikawa, T.; Yoshida, H.; Koike, K. MicroRNA-22 and microRNA-140 suppress NF-κB activity by regulating the expression of NF-κB coactivators. Biochem. Biophys. Res. Commun. 2011, 411, 826–831.
- Takata, A.; Otsuka, M.; Kudo, Y.; Kojima, K.; Maeda, S.; Tateishi, K.; Ikenoue, T.; Ijichi, H.; Hirata, Y.; Yoshida, H.; et al. Abstract 2065: DDX20 deficiency enhances NF-κB by impairing NF-κB suppressive-microRNA function and leads to hepatocarcinogenesis. Cancer Res. 2010, 70, 2065.
- Huang, Y.; Wang, C.; Li, K.; Ye, Y.; Shen, A.; Guo, L.; Chen, P.; Meng, C.; Wang, Q.; Yang, X.; et al. Death-associated protein kinase 1 suppresses hepatocellular carcinoma cell migration and invasion by upregulation of DEAD-box helicase 20. Cancer Sci. 2020, 111, 2803–2813.
- Shi, D.; Jiang, P. A Different Facet of p53 Function: Regulation of Immunity and Inflammation During Tumor Development. Front. Cell Dev. Biol. 2021, 9, 762651.
- Bordon, Y. Cell death: Tumour suppressor p53 helps phagocytes clean up. Nat. Rev. Immunol. 2015, 15, 525.
- Aylon, Y.; Oren, M. Living with p53, dying of p53. Cell 2007, 130, 597–600.
- Cooks, T.; Harris, C.C.; Oren, M. Caught in the cross fire: p53 in inflammation. Carcinogenesis 2014, 35, 1680–1690.
- Cai, Q.; Guo, Y.; Xiao, B.; Banerjee, S.; Saha, A.; Lu, J.; Glisovic, T.; Robertson, E.S. Epstein-Barr virus nuclear antigen 3C stabilizes Gemin3 to block p53-mediated apoptosis. PLoS Pathog. 2011, 7, e1002418.
- Mirzaei, S.; Zarrabi, A.; Hashemi, F.; Zabolian, A.; Saleki, H.; Ranjbar, A.; Seyed Saleh, S.H.; Bagherian, M.; Sharifzadeh, S.O.; Hushmandi, K.; et al. Regulation of Nuclear Factor-KappaB (NF-κB) signaling pathway by non-coding RNAs in cancer: Inhibiting or promoting carcinogenesis? Cancer Lett. 2021, 509, 63–80.
- Ramadass, V.; Vaiyapuri, T.; Tergaonkar, V. Small Molecule NF-κB Pathway Inhibitors in Clinic. Int. J. Mol. Sci. 2020, 21, 5164.
- Cai, W.; Xiong Chen, Z.; Rane, G.; Satendra Singh, S.; Choo, Z.; Wang, C.; Yuan, Y.; Zea Tan, T.; Arfuso, F.; Yap, C.T.; et al. Wanted DEAD/H or Alive: Helicases Winding Up in Cancers. J. Natl. Cancer Inst. 2017, 109, djw278.
- Shin, E.M.; Osato, M.; Kumar, A.P.; Tergaonkar, V. RNA helicase DP103 and TAK1: A new connection in cancer. Mol. Cell Oncol. 2015, 2, e985911.
- Bentires-Alj, M.; Barbu, V.; Fillet, M.; Chariot, A.; Relic, B.; Jacobs, N.; Gielen, J.; Merville, M.P.; Bours, V. NF-kappaB transcription factor induces drug resistance through MDR1 expression in cancer cells. Oncogene 2003, 22, 90–97.
- Shin, E.M.; Hay, H.S.; Lee, M.H.; Goh, J.N.; Tan, T.Z.; Sen, Y.P.; Lim, S.W.; Yousef, E.M.; Ong, H.T.; Thike, A.A.; et al. DEAD-box helicase DP103 defines metastatic potential of human breast cancers. J. Clin. Investig. 2014, 124, 3807–3824.
- Casey, P.; Yap, C. RNA helicase DDX20 as a surrogate marker of statin activity in invasive breast cancer. Chin. J. Pharmacol. Toxicol. 2015, S1, 84.
- Nee, G.J. Characterization of Statins-Induced DDX20 Silencing in Invasive Breast Cancers. Master’s Thesis, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, 2013.
- Chao, W. Interplay between Mevalonate and Hippo Pathways Regulates Ddx20 Transciption via Yao-Tead Complex in Triple Negative Breast Cancers. Ph.D. Thesis, National University of Singapore, Singapore, 2017.
- Pohl, S.Ö.-G. Mediation of Triple-Negative Breast Cancer Cell Fate via Cellular Redox and Wnt Signalling. Ph.D. Thesis, Curtin University, Bentley, WA, Australia, 2018.
- Zender, L.; Xue, W.; Zuber, J.; Semighini, C.P.; Krasnitz, A.; Ma, B.; Zender, P.; Kubicka, S.; Luk, J.M.; Schirmacher, P.; et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 2008, 135, 852–864.
- Chang, T.C.; Yu, D.; Lee, Y.S.; Wentzel, E.A.; Arking, D.E.; West, K.M.; Dang, C.V.; Thomas-Tikhonenko, A.; Mendell, J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008, 40, 43–50.
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233.
- Thomson, J.M.; Newman, M.; Parker, J.S.; Morin-Kensicki, E.M.; Wright, T.; Hammond, S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes. Dev. 2006, 20, 2202–2207.
- Takata, A.; Otsuka, M.; Yoshikawa, T.; Kishikawa, T.; Kudo, Y.; Goto, T.; Yoshida, H.; Koike, K. A miRNA machinery component DDX20 controls NF-κB via microRNA-140 function. Biochem. Biophys. Res. Commun. 2012, 420, 564–569.
- Ye, Y.; Spitz, M.R.; Yang, H.; Wu, X. Abstract 1174: Genetic variations in microRNA biogenesis pathway genes as susceptibility loci for lung cancer risk. Cancer Res. 2011, 71, 1174.
- Grundhoff, A.T.; Kremmer, E.; Türeci, O.; Glieden, A.; Gindorf, C.; Atz, J.; Mueller-Lantzsch, N.; Schubach, W.H.; Grässer, F.A. Characterization of DP103, a novel DEAD box protein that binds to the Epstein-Barr virus nuclear proteins EBNA2 and EBNA3C. J. Biol. Chem. 1999, 274, 19136–19144.
- Thorley-Lawson, D.A. Epstein-Barr virus: Exploiting the immune system. Nat. Rev. Immunol. 2001, 1, 75–82.
- Kawa, K. Epstein-Barr virus-associated diseases in humans. Int. J. Hematol. 2000, 71, 108–117.
- Hille, A.; Badu-Antwi, A.; Holzer, D.; Grässer, F.A. Lysine residues of Epstein-Barr virus-encoded nuclear antigen 2 do not confer secondary modifications via ubiquitin or SUMO-like proteins but modulate transcriptional activation. J. Gen. Virol. 2002, 83, 1037–1042.
- Strobl, L.J.; Höfelmayr, H.; Stein, C.; Marschall, G.; Brielmeier, M.; Laux, G.; Bornkamm, G.W.; Zimber-Strobl, U. Both Epstein-Barr Viral Nuclear Antigen 2 (EBNA2) and Activated Notch1 Transactivate Genes by Interacting with the Cellular Protein RBP-Jκ. Immunobiology 1997, 198, 299–306.
- Ling, P.D.; Rawlins, D.R.; Hayward, S.D. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc. Natl. Acad. Sci. USA 1993, 90, 9237–9241.
- Harada, S.; Kieff, E. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J. Virol. 1997, 71, 6611–6618.
- Voss, M.D.; Hille, A.; Barth, S.; Spurk, A.; Hennrich, F.; Holzer, D.; Mueller-Lantzsch, N.; Kremmer, E.; Grässer, F.A. Functional cooperation of Epstein-Barr virus nuclear antigen 2 and the survival motor neuron protein in transactivation of the viral LMP1 promoter. J. Virol. 2001, 75, 11781–11790.
- West, M.J. Structure and function of the Epstein-Barr virus transcription factor, EBNA 3C. Curr. Protein Pept. Sci. 2006, 7, 123–136.
- Subramanian, C.; Knight, J.S.; Robertson, E.S. The Epstein Barr nuclear antigen EBNA3C regulates transcription, cell transformation and cell migration. Front. Biosci. J. Virtual Libr. 2002, 7, d704–d716.
- Styles, C.T.; Paschos, K.; White, R.E.; Farrell, P.J. The Cooperative Functions of the EBNA3 Proteins Are Central to EBV Persistence and Latency. Pathogens 2018, 7, 31.
- Robert, F.; Pelletier, J. Perturbations of RNA helicases in cancer. Wiley Interdiscip. Rev. RNA 2013, 4, 333–349.
- Jäger, S.; Cimermancic, P.; Gulbahce, N.; Johnson, J.R.; McGovern, K.E.; Clarke, S.C.; Shales, M.; Mercenne, G.; Pache, L.; Li, K.; et al. Global landscape of HIV–human protein complexes. Nature 2012, 481, 365–370.
- Yang, Z.; Yang, J.; Wang, J.; Lu, X.; Jin, C.; Xie, T.; Wu, N. Identify Potential Regulators in HIV-1 Latency by Joint microRNA and mRNA Analysis. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2015, 36, 569–584.
- Greenwood, E.J.D.; Williamson, J.C.; Sienkiewicz, A.; Naamati, A.; Matheson, N.J.; Lehner, P.J. Promiscuous Targeting of Cellular Proteins by Vpr Drives Systems-Level Proteomic Remodeling in HIV-1 Infection. Cell Rep. 2019, 27, 1579–1596.e7.
