Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Lindsay Dong and Version 1 by Yundong Yuan.
Shoot branching is a complex and tightly regulated developmental process that is essential for determining plant architecture and crop yields. The outgrowth of tiller buds is a crucial step in shoot branching, and it is influenced by a variety of internal and external cues.
- bud outgrowth
- tillering
- branching
- plant hormones
1. Introduction
The plasticity exhibited by plants in their shoot development is remarkable, as it allows them to adapt to various harmful external and internal conditions in order to survive and thrive. Shoot architecture in seed plants is primarily determined by factors such as the number, position, orientation, and size of shoot branches. The regulation of shoot branching/tillering constitutes a critical survival and propagation strategy governed by a complex, sophisticated regulatory network.
Initiation of the primary shoot axis can be traced back to the shoot apical meristem (SAM), a group of mitotic cells that forms during embryogenesis. Subsequently, the derivatives of this meristem give rise to all above-ground parts of plants [1]. The SAM produces aerial organs by continuously adding growth units called phytomers, generally comprising three parts: an internode, a leaf, and an axillary meristem (AM) that emerges at the leaf axil [2].
AMs are new stem cell niches derived from the SAM during post-embryonic development. AM activity plays a vital role in generating the intricate branching patterns that contribute to a plant’s fractal architecture. Given its significant influence on shoot branching/tillering and panicle branching, the AM has been a focal point in breeding selection for improving crop production and management [3,4,5][3][4][5]. For the convenience of readers to understand the influence of crop yield, please refer to Figure 1 and Supplementary Video S1, which dynamically indicates the process of AMs’ outgrowth and their impact on grain yield.
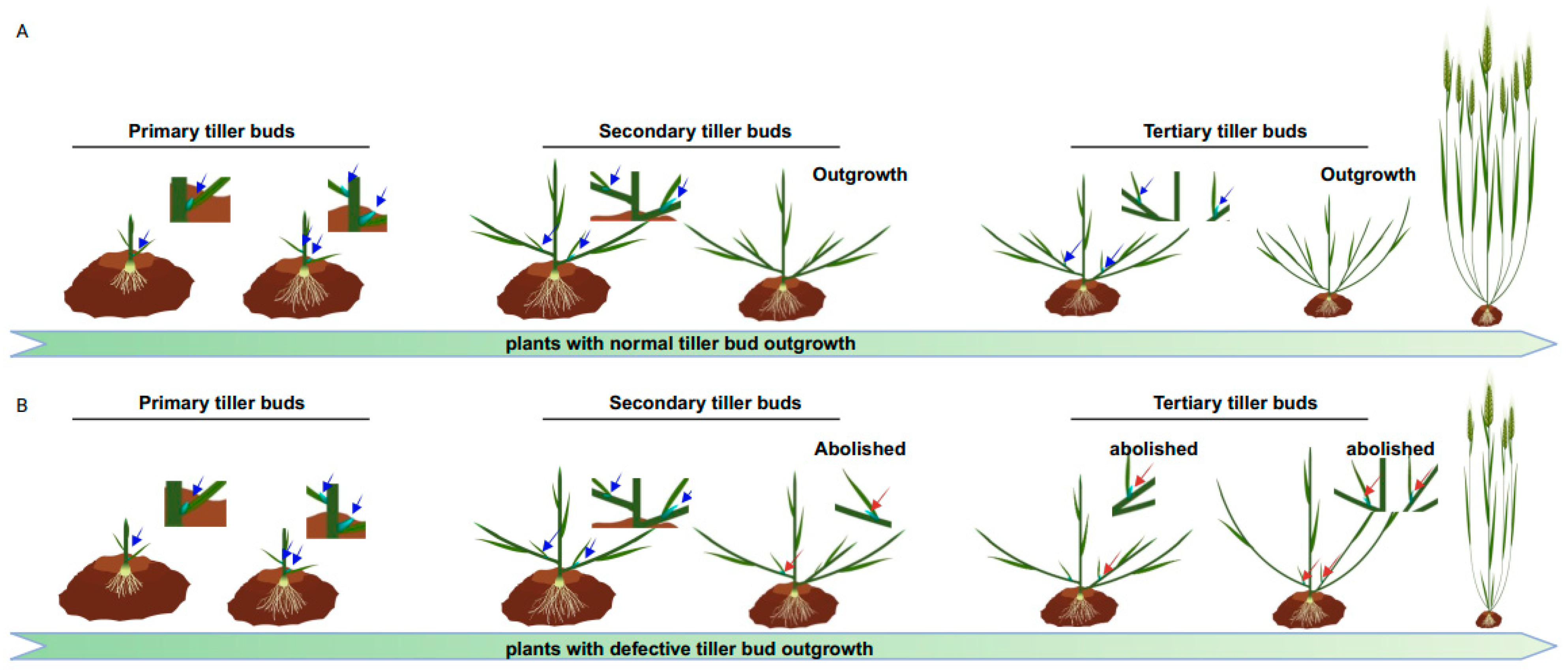
Figure 1. Illustration of the dynamic process of tiller formation and comparisons between plants with normal and defective tiller bud outgrowth. (A) portrays the successive processes of tiller bud formation and their outgrowth to generate more panicles than in (B). The primary tiller buds (arrows indicated) arise from the leaf axils of the main stem. Secondary tiller buds occur from leaf axils of the primary tillers and so on.
2. Mechanisms Regulating Shoot Branching
Once a branch/tiller bud is formed, the plant is confronted with a critical decision to stimulate bud sprouting, giving rise to a branch/tiller, or to maintain bud dormancy. Bud outgrowth typically progresses through three discernible stages: dormancy, transition, and sustained growth [24,25,26][6][7][8]. The fate of buds in the transition stage is influenced by the complex interplay of environmental and endogenous cues, ultimately determining whether buds return to dormancy or enter a sustained growth phase [27][9].
2.1. Internal Inputs Determine Bud Outgrowth
2.1.1. TB1/BRC1 Acts as a Key Integrator of Branching
The expression of TB1, encoding a non-canonical basic helix–loop–helix (bHLH) transcription factor of the TCP family, is negatively correlated with bud growth [63,64][10][11]. This TCP protein family is represented by four founding members: TB1, CYCLOIDEA (CYC), PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR1 (PCF1), and PCF2. These family members were identified by their functions in plant development or their DNA binding capacity [63,65,66,67,68][10][12][13][14][15]. The maize (Zea mays) tb1 mutant exhibits an uncontrolled proliferation of tillers, resulting in a bushy architecture reminiscent of its ancestor, teosinte [29][16]. The inhibitory effect of TB1 on bud outgrowth is spatially restricted to axillary buds as soon as they become visible [29][16]. By contrast, in teosinte, TB1 is not expressed in axillary buds, allowing axillary bud outgrowth [29][16]. The role of TB1 in suppressing axillary bud outgrowth is conserved in rice, as ectopic overexpression of its ortholog OsTB1 under the control of the actin promoter leads to reduced tillering [69][17]. Conversely, loss-of-function mutants of OsTB1, such as fine culm 1 (fc1), show increased tillering [69][17]. TB1 and its orthologs (e.g., OsTB1 or FC1 in rice, BRANCHED1 (BRC1) in Arabidopsis, PsBRC1 in pea [Pisum sativum], and SlBRC in tomato) operate in conjunction with other vital genes and plant hormones to regulate bud outgrowth [6,30,31,70][18][19][20][21]. These genes and phytohormones are discussed in subsequent sections. The coordinated action of TB1 with these factors has earned it the title “branching integrator”.2.1.2. SQUAMOSA Binding Proteins Inhibit Bud Outgrowth
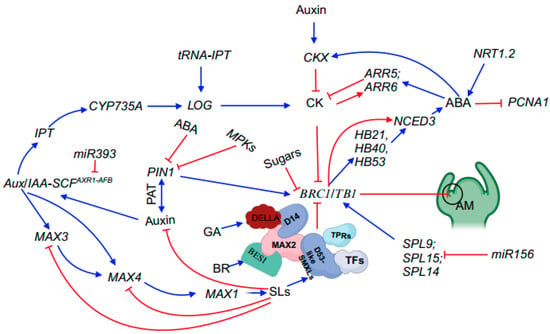
SQUAMOSA promoter binding protein-like (SPL) transcription factors, which are specific to plants, mediate various aspects of plant development, including branching [32][22]. Different members of the SPL gene family in Arabidopsis are post-transcriptionally regulated by miR156 [73][23]. Overaccumulation of miR156 leads to a considerably bushy phenotype [74,75][24][25]. Notably, double mutants of the Arabidopsis paralogs SPL9 and SPL15 exhibit an increased branching phenotype, highlighting the crucial role of miR156-targeted SPL genes in regulating shoot branching [33][26] (Figure 2).
Figure 2. Summary of phytohormones and key genes involved in shoot branching. Blue arrows represent promotion, whilst red flat-ended lines denote inhibition. In this model, BRC1/TB1 acts as an integrator to interact with other genes, such as SPL genes and phytohormones, to mediate shoot branching. Abbreviations: SL, strigolactone; ABA, abscisic acid; CK, cytokinin; GA, gibberellin; PIN1, PIN-FORMED1; SCF, Skp-Cullin-F-box; Aux/IAA, Auxin/Indole-3-Acetic Acid; IPT, ADENYLATE ISOPENTENYLTRANSFERASE; PAT, polar auxin transport; CYP735A, cytochrome P450 monooxygenase 735A; LOG, LONELY GUY; tRNA-IPT, transfer RNA isopentenyltransferase; CKX, cytokinin oxidase; ARR5 and ARR6, RESPONSE REGULATOR5 and 6; NRT1.2, nitrate transporter 1.2; PCNA1, PROLIFERATING CELL NUCLEAR ANTIGEN1; SPL9, SPL14 and SPL15, SQUAMOSA PROMOTER BINDIN PROTEIN-LIKE9,14 and 15; BRC1/TB1, BRANCHED1/TEOSINTE BRANCH 1; NCED3, 9-CIS-EPOXYCAROTENOID DIOXYGENASE3; HB21, HOMEOBOX PROTEIN 21; HB40, HOMEOBOX PROTEIN 40; HB53, HOMEOBOX PROTEIN 53; DELLA, aspartic acid–glutamic acid–leucine–leucine–alanine; D14, DWARF 14; MAX3, 2, 1, and 4, more axillary growth 3, 2, 1, and 4; TPRs, TOPLESS-RELATED PROTEINs; TFs, transcription factors; D53-like SMXLs, DWARF53-LIKE SMAX1-LIKEs; BES1, bri1-EMS-suppressor 1; AM, axillary meristem.
2.1.3. Auxin Indirectly Inhibits Sustained Bud Outgrowth
A principal function of auxin, as observed in various studies, is mediating apical dominance, as lateral bud outgrowth is inhibited by auxin. For instance, the auxin-resistant 1 (axr1) mutant of Arabidopsis exhibits a lower sensitivity to auxin than wild-type plants, resulting in weak apical dominance and an increased number of branches, due to the release of axillary buds, highlighting the role of auxin in inhibiting axillary bud outgrowth [34][27]. Moreover, apical dominance largely depends on polar auxin transport (PAT) mediated by PIN-FORMED (PIN) proteins in the stem [76][28]. Studies involving RNA interference (RNAi)-mediated knockdown and overexpression of OsPIN1 have demonstrated the negative effect of OsPIN1 on tillering in rice [36][29]. In addition to the crucial role of PAT in regulating branching/tillering, the auxin signaling pathway also influences shoot branching/tillering via SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Auxin receptors, including TRANSPORT INHIBITOR RESPONSE 1 (TIR1) and closely related family numbers (AUXIN SIGNALING F-BOX [AFB]) [9[30][31][32],40,41], bind to auxin to stabilize the interactions between TIR1/AFBs and members of the Aux/IAA (Auxin/INDOLE-3-ACETIC ACID INDUCIBLE) family of transcriptional repressors [77][33]. Their interaction with TIR1/AFBs leads to the degradation of Aux/IAA, permitting auxin-mediated upregulation of transcription [78,79,80][34][35][36]. By contrast, loss of function of IAA12 in Arabidopsis leads to auxin-resistant stabilization of the SCF complex and, thus, constitutive suppression of target auxin-upregulated genes [41][32], resulting in a bushy phenotype. Overexpressing OsMIR393, whose mature miRNA product OsmiR393 targets and downregulates the transcripts of the auxin receptor genes OsTIR1 and OsAFB2, leads to increased tiller production [42][37].2.1.4. Strigolactones Have an Inhibitory Effect on Bud Outgrowth
SLs are a collection of carotenoid-derived lactones secreted by plants. These phytohormones are primarily known for their roles as rhizosphere signals used by root-parasitic plants to detect their hosts [96][38] and as cues for mycorrhizal fungi to form symbiotic associations [97][39]. Importantly, SLs also inhibit the outgrowth of axillary buds. This inhibitory effect was initially observed in ramosus (rms) mutants in pea, which exhibit excessive branching, as well as decreased apical dominance (dad) mutants in petunia (Petunia hybrida) and more axillary growth (max) mutants in Arabidopsis [45,98,99,100,101,102,103,104][40][41][42][43][44][45][46][47]. SL biosynthesis involves several enzymes. DWARF27 (D27) is responsible for isomerizing all-trans-β-carotene at the C-9 position to form 9-cis-carotene [12,43][48][49]. Carotenoid cleavage dioxygenase 7 (CCD7, also named MAX3) and CCD8 (also named MAX4) then cleave 9-cis-carotene to produce carlactone, a key endogenous SL precursor [44,45][40][50]. The conversion of carlactone to SLs is catalyzed by MAX1, an Arabidopsis cytochrome P450 that acts downstream of MAX4 and MAX3 [46][51]. MAX1 converts carlactone to carlactonoic acid, which is further converted to methyl carlactonoate, an SL-like compound (Figure 2) [105][52]. SLs are perceived by MAX2, an ortholog of D3 from rice, which plays a crucial role in regulating plant branching. Disruption of MAX2 leads to a bushy phenotype in Arabidopsis [46][51]. Moreover, the perception and signaling roles of SLs in rice require their interaction with D14, a putative SL receptor. D14 interacts with D3, an F-box protein of the SCF E3 ubiquitin ligase complex, to form an SL-induced D14-D3 complex [47][53]. This complex targets proteins for ubiquitination and degradation, resulting in changes in plant branching/tillering [48][54] (Figure 2).The dominant d53 mutant in rice is characterized by a high-tillering and dwarf phenotype and is resistant to the exogenous application of GR24, a synthetic SL. D53 is targeted for SL-dependent degradation by the SCFD3 ubiquitination complex [106][55]. In addition, SL treatment results in D53 degradation via the ubiquitin–proteasome system in a D14- and D3-dependent manner [48][54]. D53 interacts with members of the TOPLESS-RELATED PROTEIN (TPR) family of transcriptional co-repressors, which may suppress the activities of their downstream transcription factors (Figure 2) [47,107][53][56].
2.1.5. Other Phytohormones Regulate Tillering/Branching
The inhibitory role of auxin in bud outgrowth is exerted indirectly within buds, as apically derived auxin is not transported into buds [108][57] and exogenous auxin directly supplied to buds fails to prevent their growth [109][58]. Cytokinins (CKs) are believed to be crucial in relaying the auxin signal into buds and promoting axillary branching. CKs are an important class of phytohormones that participate in various aspects of plant development, including organ formation, apical dominance, and leaf senescence [110][59]. Therefore, CKs facilitate the growth and development of axillary buds by serving as a second messenger for the auxin signal. Several studies have demonstrated that CKs can promote the outgrowth of buds that would otherwise remain inhibited and that CK levels in or near the bud are well correlated with bud fate [50,111][60][61]. For example, in chickpea (Cicer arietinum L.) plants, CK levels dramatically increased in axillary buds within 24 h of shoot decapitation [112][62]. Elevated levels of CKs, as achieved by overexpressing ISOPENTENYL TRANSFERASE (IPT) (encoding a key enzyme in CK biosynthesis), lead to reduced apical dominance (Figure 2) [50][60]. Continuous treatment of pea plants with synthetic CKs overcomes the inhibition of lateral bud release, turning these into dominant organs [113][63]. In addition to their biosynthesis, the metabolism of CKs also determines endogenous cytokinin levels. Cytokinin oxidase (CKX) is the enzyme responsible for inactivating CKs by irreversibly degrading active CKs, thereby regulating endogenous levels of active CKs [116,117][64][65]. Various CKXs, such as PsCKX2 in pea, predominantly regulate CK levels in the stem [52][66]. While abscisic acid (ABA) is predominantly recognized for its roles in seed dormancy, growth inhibition, and stress responses, emerging evidence indicates that this phytohormone also influences plant branching/tillering. Mutations in key genes involved in ABA biosynthesis, such as 9-CIS-EPOXYCAROTENOID DIOXYGENASE3 (NCED3) and ABA DEFICIENT2 (ABA2), enhance bud outgrowth [53,54][67][68]. Notably, ABA levels are elevated in buds with delayed outgrowth but are reduced in elongated buds [54][68]. Exogenous application of ABA partially suppresses branch elongation, suggesting that ABA functions downstream or independently of genes responsible for bud growth [54][68]. Unlike the IAA biosynthesis gene TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1) and the auxin transporter gene PIN1, exogenously supplied ABA does not affect BRC1 expression, confirming the downstream role of ABA in regulating BRC1. Furthermore, BRC1 binds to the promoter of and positively regulates the transcription of three genes encoding related homeodomain leucine zipper proteins (HD-ZIP): HOMEOBOX PROTEIN 21 (HB21), HB40, and HB53. Together with BRC1, these three proteins promote NCED3 expression, resulting in ABA accumulation and triggering a phytohormonal response, thereby suppressing bud development [14][69] (Figure 2). Gibberellins (GAs) are renowned for their ability to regulate internode elongation. Dwarfism is often associated with an increase in shoot branching. GA-deficient mutants in Arabidopsis, rice, and pea exhibit higher levels of branching/tillering than their wild-type counterparts [10,118][70][71]. Brassinosteroids (BRs) are important plant hormones that regulate various developmental processes, including stem elongation, leaf development, senescence, and branching [119][72]. A series of BR-deficient mutants have been used to elucidate the function of BR. For instance, reduced expression of Brassinazole Resistant 1 (OsBZR1) in rice leads to a dwarf phenotype with erected leaves and reduced BR sensitivity [60][73]. Dwarf and Low Tillering (DLT), encoding a GRAS family protein, is another BR-related gene. Disruption of DLT results in a semi-dwarf mutant with fewer tillers and decreased BR responses. The promoter of DLT can be targeted by OsBZR1 [61][74]. GLYCOGEN SYNTHASE KINASE 2 (GSK2) encodes a conserved glycogen synthase kinase 3-like kinase. Gain-of-function mutations within the GSK2 coding sequencing or its overexpression suppress BR signaling, leading to plants with phenotypes resembling BR-deficient mutants [120][75]. The reduced leaf angle 1 (rla1) mutant encodes a transcription factor containing an APETALA2 (AP2) DNA binding domain that is required for OsBZR1 function [62][76]. RLA also can interact with GSK2 [62][76]. A BR-defective rice mutant displays reduced branching like dlt [61][74], while in Arabidopsis, the bri1-EMS-suppressor 1 (bes1) mutant displays a highly branched phenotype. In contrast, BES1-RNAi lines have fewer branches than the wild type [55,119][72][77]. Interestingly, the dominant bes1-D mutant does not respond to GR24 treatment, and BES1 can interact with MAX2 and act as its substrate for degradation, which is regulated by SLs [15][78]. Therefore, SL and BR signaling pathways converge on the same transcription factor BES1, an Arabidopsis homolog of OsBZR1, to control branching [15][78]. However, other components upstream of AtBES1 in the BR signaling pathway do not alter branching in Arabidopsis [15][78].2.1.6. Phytohormones Interact Influencing Bud Outgrowth
The regulation of axillary bud outgrowth involves a complex network of phytohormones. Plant hormones, including auxin, CKs, and SLs, play central roles in bud outgrowth. Furthermore, plant hormones can mutually affect each other, ultimately regulating branching/tillering. Auxin is an indispensable player in plant architecture, particularly branching, and a central component of interacting networks that regulate branching. Auxin exerts its role indirectly in buds [85][79], whereas SLs are direct components that act on buds via auxin to inhibit bud outgrowth. Auxin inhibits bud outgrowth by regulating SL biosynthesis, as observed in Arabidopsis and rice [92,121,122][80][81][82]. For example, auxin that originates in the shoot apex modulates SL levels in pea by maintaining RMS1 and RMS5 transcript abundance [123,124][83][84]. The iaa12 mutant exhibits increased numbers of branches and shows reduced expression of the SL biosynthesis genes MAX3 and MAX4 [92][80], while MAX3 and MAX4 transcription is mediated by auxin [125][85]. Conversely, Arabidopsis mutants defective in SL biosynthesis (such as max4) exhibit increased branching and resistance to auxin [45][40]. SLs also inversely affect polar auxin transport by limiting the accumulation of the auxin efflux carrier PIN1 in cells involved in polar auxin transport [87][86]. The Arabidopsis max mutants, with defects in the SL pathway, show enhanced polar PIN accumulation and auxin transport [127,128,129][87][88][89]. Auxin and CKs play antagonistic roles in regulating bud outgrowth. Auxin inhibits AM outgrowth, whereas CKs counteract auxin activity in Arabidopsis by promoting bud activation [20][90]. The inhibitory effect of auxin is probably mediated, at least in part, by its ability to reduce both CK export from roots and CK biosynthesis locally at the node [130,131][91][92]. Stem girdling in pea, which prevents polar auxin transport via a mechanism similar to decapitation, increases the expression of CK biosynthesis genes and promotes the growth of buds below the girdling site [115][93]. Decapitated bean (Phaseolus vulgaris L.) plants (with a decrease or loss of polar auxin transport) have higher CK concentrations in xylem exudates than control plants. However, applying auxin to the shoots of decapitated plants eliminates the effect of shoot-tip removal on CK concentration, further supporting the antagonistic relationship between auxin and CKs [130][91]. Auxin inhibits expression of the CK biosynthesis gene IPT in the stem [111,115,130,132][61][91][93][94].2.1.7. Sugars Play an Essential Role in Bud Release
Sugars such as sucrose serve not only as a carbon source for plant metabolism but also as essential signaling compounds [137,138,139][95][96][97]. Bud outgrowth responds to limiting nutrients and resources, and thus sugars have been attracting increasing interest. Axillary buds are often maintained in a dormant state or their growth is suppressed by the growing shoot apex long after their initial formation. Intriguingly, changes in sugar availability can facilitate the first visible growth of buds, a phenomenon known as axillary bud release [16][98]. The term “apical dominance” is commonly used to describe shoot branching, referring to the role of the shoot tip in preventing the growth of the axillary buds below it [140][99]. However, in several plant species, auxin supplementation to the decapitated stump, even at high levels, fails to fully restore apical dominance [109,141][58][100]. Enhancing the sugar supply alone is sufficient for bud release. Plants employ various mechanisms to maintain apical dominance, one of which is limiting the sugar supply to axillary buds [16][98]. This effect can be observed in the wheat tiller inhibition (tin) mutant, whose reduced tillering is associated with a decreased sucrose content in axillary buds [143][101]. Likewise, reduced tiller formation in the rice monoculm 2 (moc2) mutant is attributed to a disruption in fructose-1,6-bisphosphatase, an enzyme involved in sucrose biosynthesis, resulting in a decline in the sucrose supply [56][102]. The requirement for sugars for bud outgrowth has been demonstrated in rose (Rosa hybrida), where sugar is required for triggering bud outgrowth in single nodes cultivated in vitro [88,144][103][104]. Sucrose can also modulate the dynamics of bud outgrowth in a concentration-dependent manner, especially during the transition phase between bud release and sustained bud elongation [88][103]. Besides the roles of sugars as nutrients, an effect of sugars on phytohormone homeostasis has been demonstrated in single nodes of R. hybrida [88][103]. For instance, sucrose stimulates CK biosynthesis in bud-bearing stem segments by upregulating the expression of two CK biosynthesis-related genes [88][103]. Sucrose can also modulate auxin metabolism, as treatment with sucrose or its non-metabolizable analogs increases auxin levels in R. hybrida buds in a concentration-dependent manner [88][103].2.2. Effects of Environmental Inputs on Bud Outgrowth
Bud outgrowth and the transition to dormancy are tightly regulated by various environmental factors, including photoperiod, light intensity, nutrient availability, and stress conditions [26,147][8][105]. Notably, ABA, a pivotal phytohormone involved in regulating bud outgrowth, strongly accumulates under stressful conditions such as osmotic stress [148][106]. Interestingly, ABA levels are elevated in buds with delayed outgrowth but are reduced in elongated buds [54][68]. This dynamic modulation of ABA levels underscores the remarkable ability of plants to adjust their branching capability in response to diverse environmental cues. Such adaptability represents a successful evolutionary trait that has evolved to accommodate the sessile nature of plants.2.2.1. Light Plays a Critical Role in Bud Outgrowth
Light intensity is pivotal for regulating bud outgrowth across numerous plant species. For instance, low-intensity light inhibits tillering in wheat [163][107]. By contrast, high-intensity light stimulates branching, as observed in Rosa species [164][108]. Photoperiod also has a significant influence on the distribution of bud outgrowth along the plant stem. For example, under short-day conditions, the formation of basal branches is enhanced in pea. Bud outgrowth in the upper nodes often coincides with the onset of flowering and may also be controlled by photoperiod [165,166][109][110]. At high density, shade also regulates bud dormancy in cultivated plants [167][111]. When plants intercept incident light, the light intensity decreases, preferentially in the red part of the light spectrum. Shade is, therefore, characterized by a reduction in the red (R) to far-red (FR) light ratio (R:FR) due to R light absorption and FR reflection by leaves [168][112]. This decrease in R:FR serves as a signal of shade or competition for light, prompting plants to respond by inhibiting axillary bud outgrowth, elongating their stature, and accelerating flowering to evade the detrimental consequences of shading. This suite of responses, known as shade avoidance syndrome, is mediated by the R- and FR-absorbing photoreceptor phytochrome B (PHYB) [169][113]. In densely grown sorghum (Sorghum bicolor) plants experiencing shade, inhibition of bud outgrowth due to an enriched FR-light environment is associated with activation of the TB1-like gene SbTB1 in buds [13,149][114][115].2.2.2. Impacts of Nutrients on Bud Outgrowth
Nitrogen (N) is an essential macronutrient that dominates plant growth and plant productivity [170,171][116][117]. N limitation can significantly limit the tiller numbers in rice [170][116]. Furthermore, high branching ability is positively correlated with the capacity for N uptake. Several N transporters have been identified that regulate shoot branching in rice. One such transporter is peptide transporters family 7.7 (OsNPF7.7), whose increased abundance facilitates the influx of NO3− and NH4+, thereby promoting the outgrowth of axillary buds [57][118]. Indica NADH/NADPH-DEPENDENT NITRATE REDUCTASE 2 (OsNR2) promotes NO3− uptake through interaction with OsNRT1.1B, a low-affinity NO3− transport gene, and increases effective tillers in japonica rice. Similarly, the japonica allele of OsNR2 also promotes tillering but not to the extent observed in indica OsNR2-overexpression lines [150,172][119][120]. The rice APETALA2-domain transcription factor encoded by a NITROGEN MEDIATED TILLER GROWTH RESPONSE 5 (NGR5) allele is upregulated under conditions of increased nitrogen availability. NGR5 interacts with a component of the polycomb repressive complex 2 (PRC2) to regulate the expression of D14 and OsSPL14 by mediating levels of histone methylation (H3K27me3) modification, thereby regulating rice tillering [151,172][120][121]. Overexpression of DENSE AND ERECT PANICLE1 (OsDEP1) can increase tiller numbers under high N supply [152][122]. Given that N availability influences CK and SL levels, it is plausible that N functions as a second messenger to mediate branching/tillering [174,175,176][123][124][125]. In several species, limited N availability promotes SL production, subsequently inhibiting branching/tillering. N limitation also leads to a reduction in CK production. N fertilization can suppress the expression of SL biosynthesis genes [171,177][117][126]. Phosphorus (Pi) is an essential macronutrient for plant growth and metabolism. However, the availability of Pi in soils is often low due to chemical fixation and poor diffusion, resulting in low-Pi environments that can limit plant development and processes like tillering [178][127]. For example, Pi deficiency has been shown to reduce tiller numbers in rice [179][128], with the transcription factor Oryza sativa PHOSPHATE STARVATION RESPONSE2 (OsPHR2) implicated in the repression of tillering under low Pi conditions [155][129]. Pi deficiency also induced SL biosynthesis by increasing transcription of SL biosynthetic genes like the β-carotene cis-trans isomerase DWARF27 (D27), the carotenoid cleavage dioxygenase 7 (CCD7)/D17, and CCD8/D10 [2[2][128][130],179,180], which play a key role in regulating tillering, as described above. Potassium (K+) is the most abundant cation in plants and an essential macronutrient [157,181][131][132]. Adequate K+ availability can increase tillering in plants, as evidenced by enhanced tillering in rice overexpressing Oryza sativa High-Affinity K+ Transporter 5 (OsHAK5). In contrast, knockout of OsHAK5 reduces tillers in rice [157][131], producing a phenotype resembling loss-of-function mutants of the auxin transporter OsABCB14 [158][133], implying a potential interaction between K+ and auxin.2.2.3. Water Availability Influences Bud Outgrowth
Tillering/branching processes are susceptible to drought stress, one of the most limiting factors affecting agricultural yields. Optimal water availability is critical for normal plant growth and development, including tillering. Tolerance to this stress is multigenic and complex in nature. Drought stress triggers specific alterations of gene-expression patterns in plant tissues [182][134]. For instance, the drought-inducible microRNA miR393 was shown to be upregulated in Arabidopsis [183][135], and miR393 also regulates tiller number increases in rice by modulating auxin signaling through auxin receptor genes like OsAUX1 and OsTIR1.2.2.4. Effects of Temperature on Tillering
Temperature is a critical factor influencing tillering in crops [184][136]. High temperatures caused by extreme weather events can reduce tiller numbers, as evidenced in B. distachyon. Tiller numbers declined linearly in B. distachyon from 24 to 36 °C at a rate of approximately one tiller for every 1.7 °C increase in temperature [185][137]. Genes involved in heat stress play an important role in tillering. For instance, in rice, miR159 is downregulated by heat stress, and its overexpression increases heat sensitivity and significantly reduces tillering [186][138]. At the other extreme, chilling also detrimentally impacts tillering. Chilling tolerance is a complex agronomic trait governed by intricate genetic networks and signal transduction cascades. Mechanistic insights into cold-stress effects on tillering are emerging. For example, overexpression of OsMADS57 maintains rice tiller growth under chilling stress. OsMADS57 directly binds and activates the defense gene OsWRKY94 for cold-stress responses while suppressing its activity under normal temperatures [187][139].2.2.5. Biotic Stresses Impact Tiller Bud Outgrowth
Biotic stress affects plant development, including tillering, fundamentally disrupting and depriving plants of the nutrients they rely on for survival. More specifically, biotic stresses caused by plant pathogens, insect pests, and parasitic organisms can impair growth and developmental processes such as tillering or branching (Figure 3). These biotic agents injure plant tissues both directly through feeding/infection and indirectly by inhibiting the uptake and utilization of water, nutrients, and photoassimilates required for plant growth. Pathogens, insects, and parasites disrupt key physiological processes like metabolism, resource allocation, and energy balance, ultimately reducing the plant’s capacity for producing new tillers or branches.References
- Bowman, J.L.; Eshed, Y. Formation and maintenance of the shoot apical meristem. Trends Plant Sci. 2000, 5, 110–115.
- Wang, B.; Smith, S.M.; Li, J. Genetic Regulation of Shoot Architecture. Annu. Rev. Plant Biol. 2018, 69, 437–468.
- Springer, N. Shaping a better rice plant. Nat. Genet. 2010, 42, 475–476.
- Wang, Y.; Li, J. Molecular Basis of Plant Architecture. Annu. Rev. Plant Biol. 2008, 59, 253–279.
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544.
- Stafstrom, J.P.; Sussex, I.M. Expression of a Ribosomal Protein Gene in Axillary Buds of Pea Seedlings. Plant Physiol. 1992, 100, 1494–1502.
- Devitt, M.L.; Stafstrom, J.P. Cell cycle regulation during growth-dormancy cycles in pea axillary buds. Plant Mol. Biol. 1995, 29, 255–265.
- Dun, E.A.; Ferguson, B.J.; Beveridge, C.A. Apical dominance and shoot branching. Divergent opinions or divergent mechanisms? Plant Physiol. 2006, 142, 812–819.
- Waldie, T.; Hayward, A.; Beveridge, C.A. Axillary bud outgrowth in herbaceous shoots: How do strigolactones fit into the picture? Plant Mol. Biol. 2010, 73, 27–36.
- Doebley, J.; Stec, A.; Hubbard, L. The evolution of apical dominance in maize. Nature 1997, 386, 485–488.
- Cubas, P.; Lauter, N.; Doebley, J.; Coen, E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 1999, 18, 215–222.
- Doebley, J.; Stec, A.; Gustus, C. teosinte branched1 and the origin of maize: Evidence for epistasis and the evolution of dominance. Genetics 1995, 141, 333–346.
- Doebley, J. The genetics of maize evolution. Annu. Rev. Genet. 2004, 38, 37–59.
- Luo, D.; Carpenter, R.; Vincent, C.; Copsey, L.; Coen, E. Origin of floral asymmetry in Antirrhinum. Nature 1996, 383, 794–799.
- Kosugi, S.; Ohashi, Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 1997, 9, 1607–1619.
- Hubbard, L.; McSteen, P.; Doebley, J.; Hake, S. Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 2002, 162, 1927–1935.
- Takeda, T.; Suwa, Y.; Suzuki, M.; Kitano, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M.; Ueguchi, C. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 2003, 33, 513–520.
- Aguilar-Martínez, J.A.; Poza-Carrión, C.; Cubas, P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 2007, 19, 458–472.
- Martín-Trillo, M.; Grandío, E.G.; Serra, F.; Marcel, F.; Rodríguez-Buey, M.L.; Schmitz, G.; Theres, K.; Bendahmane, A.; Dopazo, H.; Cubas, P. Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. 2011, 67, 701–714.
- Braun, N.; de Saint Germain, A.; Pillot, J.-P.; Boutet-Mercey, S.; Dalmais, M.; Antoniadi, I.; Li, X.; Maia-Grondard, A.; Le Signor, C.; Bouteiller, N. The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol. 2012, 158, 225–238.
- Minakuchi, K.; Kameoka, H.; Yasuno, N.; Umehara, M.; Luo, L.; Kobayashi, K.; Hanada, A.; Ueno, K.; Asami, T.; Yamaguchi, S.; et al. FINE CULM1 (FC1) Works Downstream of Strigolactones to Inhibit the Outgrowth of Axillary Buds in Rice. Plant Cell Physiol. 2010, 51, 1127–1135.
- Rameau, C.; Bertheloot, J.; Leduc, N.; Andrieu, B.; Foucher, F.; Sakr, S. Multiple pathways regulate shoot branching. Front. Plant Sci. 2015, 5, 741–755.
- Rhoades, M.W.; Reinhart, B.J.; Lim, L.P.; Burge, C.B.; Bartel, B.; Bartel, D.P. Prediction of Plant MicroRNA Targets. Cell 2002, 110, 513–520.
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific Effects of MicroRNAs on the Plant Transcriptome. Dev. Cell 2005, 8, 517–527.
- Wei, S.; Gruber, M.Y.; Yu, B.; Gao, M.-J.; Khachatourians, G.G.; Hegedus, D.D.; Parkin, I.A.; Hannoufa, A. Arabidopsis mutant sk156 reveals complex regulation of SPL15 in a miR156-controlled gene network. BMC Plant Biol. 2012, 12, 169.
- Schwarz, S.; Grande, A.V.; Bujdoso, N.; Saedler, H.; Huijser, P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 2008, 67, 183–195.
- Lincoln, C.; Britton, J.H.; Estelle, M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 1990, 2, 1071–1080.
- Matthes, M.S.; Best, N.B.; Robil, J.M.; Malcomber, S.; Gallavotti, A.; McSteen, P. Auxin EvoDevo: Conservation and Diversification of Genes Regulating Auxin Biosynthesis, Transport, and Signaling. Mol. Plant 2019, 12, 298–320.
- Xu, M.; Zhu, L.; Shou, H.; Wu, P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 2005, 46, 1674–1681.
- Dharmasiri, N.; Dharmasiri, S.; Estelle, M. The F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 441–445.
- Kepinski, S.; Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 446–451.
- Dharmasiri, N.; Dharmasiri, S.; Weijers, D.; Lechner, E.; Yamada, M.; Hobbie, L.; Ehrismann, J.S.; Jürgens, G.; Estelle, M. Plant Development Is Regulated by a Family of Auxin Receptor F Box Proteins. Dev. Cell 2005, 9, 109–119.
- Tan, X.; Calderon-Villalobos, L.I.A.; Sharon, M.; Zheng, C.; Robinson, C.V.; Estelle, M.; Zheng, N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 2007, 446, 640–645.
- Zenser, N.; Ellsmore, A.; Leasure, C.; Callis, J. Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 2001, 98, 11795–11800.
- Tiwari, S.B.; Wang, X.-J.; Hagen, G.; Guilfoyle, T.J. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 2001, 13, 2809–2822.
- Gray, W.M.; Kepinski, S.; Rouse, D.; Leyser, O.; Estelle, M. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 2001, 414, 271–276.
- Xia, K.; Wang, R.; Ou, X.; Fang, Z.; Tian, C.; Duan, J.; Wang, Y.; Zhang, M. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE 2012, 7, e30039.
- Cook, C.; Whichard, L.P.; Turner, B.; Wall, M.E.; Egley, G.H. Germination of witchweed (Striga lutea Lour.): Isolation and properties of a potent stimulant. Science 1966, 154, 1189–1190.
- Akiyama, K.; Matsuzaki, K.-I.; Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005, 435, 824–827.
- Sorefan, K. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 2003, 17, 1469–1474.
- Napoli, C.; Ruehle, J. New mutations affecting meristem growth and potential in Petunia hybrida Vilm. J. Hered. 1996, 87, 371–377.
- Beveridge, C.A.; Symons, G.M.; Murfet, I.C.; Ross, J.J.; Rameau, C. The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissible signal (s). Plant Physiol. 1997, 115, 1251.
- Beveridge, C.A. Long-distance signalling and a mutational analysis of branching in pea. Plant Growth Regul. 2000, 32, 193–203.
- Beveridge, C.A. Axillary bud outgrowth: Sending a message. Curr. Opin. Plant Biol. 2006, 9, 35–40.
- Turnbull, C.G.N.; Booker, J.P.; Leyser, H.M.O. Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J. 2002, 32, 255–262.
- Foo, E.; Turnbull, C.G.N.; Beveridge, C.A. Long-Distance Signaling and the Control of Branching in therms1 Mutant of Pea. Plant Physiol. 2001, 126, 203–209.
- Morris, S.E.; Turnbull, C.G.; Murfet, I.C.; Beveridge, C.A. Mutational analysis of branching in pea. Evidence that Rms1 and Rms5 regulate the same novel signal. Plant Physiol. 2001, 126, 1205–1213.
- Lin, H.; Wang, R.; Qian, Q.; Yan, M.; Meng, X.; Fu, Z.; Yan, C.; Jiang, B.; Su, Z.; Li, J.; et al. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 2009, 21, 1512–1525.
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.; Beyer, P.; Al-Babili, S. The Path from β-Carotene to Carlactone, a Strigolactone-Like Plant Hormone. Science 2012, 335, 1348–1351.
- Booker, J.; Auldridge, M.; Wills, S.; McCarty, D.; Klee, H.; Leyser, O. MAX3/CCD7 Is a Carotenoid Cleavage Dioxygenase Required for the Synthesis of a Novel Plant Signaling Molecule. Curr. Biol. 2004, 14, 1232–1238.
- Booker, J.; Sieberer, T.; Wright, W.; Williamson, L.; Willett, B.; Stirnberg, P.; Turnbull, C.; Srinivasan, M.; Goddard, P.; Leyser, O. MAX1 Encodes a Cytochrome P450 Family Member that Acts Downstream of MAX3/4 to Produce a Carotenoid-Derived Branch-Inhibiting Hormone. Dev. Cell 2005, 8, 443–449.
- Abe, S.; Sado, A.; Tanaka, K.; Kisugi, T.; Asami, K.; Ota, S.; Kim, H.I.; Yoneyama, K.; Xie, X.; Ohnishi, T.; et al. Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc. Natl. Acad. Sci. 2014, 111, 18084–18089.
- Smith, S.M.; Li, J. Signalling and responses to strigolactones and karrikins. Curr. Opin. Plant Biol. 2014, 21, 23–29.
- Wang, L.; Wang, B.; Jiang, L.; Liu, X.; Li, X.; Lu, Z.; Meng, X.; Wang, Y.; Smith, S.M.; Li, J. Strigolactone Signaling in Arabidopsis Regulates Shoot Development by Targeting D53-Like SMXL Repressor Proteins for Ubiquitination and Degradation. Plant Cell 2015, 27, 3128–3142.
- Jiang, L.; Liu, X.; Xiong, G.; Liu, H.; Chen, F.; Wang, L.; Meng, X.; Liu, G.; Yu, H.; Yuan, Y.; et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 2013, 504, 401–405.
- Xiong, G.; Wang, Y.; Li, J. Action of strigolactones in plants. Enzymes 2014, 35, 57–84.
- Morris, D.A. Transport of exogenous auxin in two-branched dwarf pea seedlings (Pisum sativum L.). Planta 1977, 136, 91–96.
- CLINE, M.G. Exogenous auxin effects on lateral bud outgrowth in decapitated shoots. Ann. Bot. 1996, 78, 255–266.
- El-Showk, S.; Ruonala, R.; Helariutta, Y. Crossing paths: Cytokinin signalling and crosstalk. Development 2013, 140, 1373–1383.
- Medford, J.I.; Horgan, R.; El-Sawi, Z.; Klee, H.J. Alterations of Endogenous Cytokinins in Transgenic Plants Using a Chimeric Isopentenyl Transferase Gene. Plant Cell 1989, 1, 403–413.
- Tanaka, M.; Takei, K.; Kojima, M.; Sakakibara, H.; Mori, H. Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 2006, 45, 1028–1036.
- Turnbull, C.G.; Raymond, M.A.; Dodd, I.C.; Morris, S.E. Rapid increases in cytokinin concentration in lateral buds of chickpea (Cicer arietinum L.) during release of apical dominance. Planta 1997, 202, 271–276.
- Li, C.; Bangerth, F. The possible role of cytokinins, ethylene and indoleacetic acid in apical dominance. In Progress in Plant Growth Regulation. Current Plant Science and Biotechnology in Agriculture; Springer: Dordrecht, The Netherlands, 1992; pp. 431–436.
- Jones, R.J.; Schreiber, B. Role and function of cytokinin oxidase in plants. Plant Growth Regul. 1997, 23, 123–134.
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487–10492.
- Shimizu-Sato, S.; Tanaka, M.; Mori, H. Auxin–cytokinin interactions in the control of shoot branching. Plant Mol. Biol. 2009, 69, 429–435.
- Reddy, S.K.; Holalu, S.V.; Casal, J.J.; Finlayson, S.A. Abscisic acid regulates axillary bud outgrowth responses to the ratio of red to far-red light. Plant Physiol. 2013, 163, 1047–1058.
- Yao, C.; Finlayson, S.A. Abscisic Acid Is a General Negative Regulator of Arabidopsis Axillary Bud Growth. Plant Physiol. 2015, 169, 611–626.
- González-Grandío, E.; Pajoro, A.; Franco-Zorrilla, J.M.; Tarancón, C.; Immink, R.G.H.; Cubas, P. Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP/cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. USA 2017, 114, E245–E254.
- Lo, S.-F.; Yang, S.-Y.; Chen, K.-T.; Hsing, Y.-I.; Zeevaart, J.A.D.; Chen, L.-J.; Yu, S.-M. A Novel Class of Gibberellin 2-Oxidases Control Semidwarfism, Tillering, and Root Development in Rice. Plant Cell 2008, 20, 2603–2618.
- Silverstone, A.L.; Mak, P.Y.A.; Martinez, E.C.; Sun, T.-P. The New RGA Locus Encodes a Negative Regulator of Gibberellin Response in Arabidopsis thaliana. Genetics 1997, 146, 1087–1099.
- Yin, Y.; Wang, Z.-Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 Accumulates in the Nucleus in Response to Brassinosteroids to Regulate Gene Expression and Promote Stem Elongation. Cell 2002, 109, 181–191.
- Bai, M.Y.; Zhang, L.Y.; Gampala, S.S.; Zhu, S.W.; Song, W.Y.; Chong, K.; Wang, Z.Y. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 2007, 104, 13839–13844.
- Tong, H.; Jin, Y.; Liu, W.; Li, F.; Fang, J.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009, 58, 803–816.
- Li, J.; Nam, K.H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 2002, 295, 1299–1301.
- Qiao, S.; Sun, S.; Wang, L.; Wu, Z.; Li, C.; Li, X.; Wang, T.; Leng, L.; Tian, W.; Lu, T.; et al. The RLA1/SMOS1 Transcription Factor Functions with OsBZR1 to Regulate Brassinosteroid Signaling and Rice Architecture. Plant Cell 2017, 29, 292–309.
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A New Class of Transcription Factors Mediates Brassinosteroid-Regulated Gene Expression in Arabidopsis. Cell 2005, 120, 249–259.
- Wang, Y.; Sun, S.; Zhu, W.; Jia, K.; Yang, H.; Wang, X. Strigolactone/MAX2-Induced Degradation of Brassinosteroid Transcriptional Effector BES1 Regulates Shoot Branching. Dev. Cell 2013, 27, 681–688.
- Prasad, T.; Li, X.; Abdel-Rahman, A.; Hosokawa, Z.; Cloud, N.; Lamotte, C.; Cline, M. Does auxin play a role in the release of apical dominance by shoot inversion in Ipomoea nil? Ann. Bot. 1993, 71, 223–229.
- Hayward, A.; Stirnberg, P.; Beveridge, C.; Leyser, O. Interactions between Auxin and Strigolactone in Shoot Branching Control. Plant Physiol. 2009, 151, 400–412.
- Arite, T.; Iwata, H.; Ohshima, K.; Maekawa, M.; Nakajima, M.; Kojima, M.; Sakakibara, H.; Kyozuka, J. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 2007, 51, 1019–1029.
- Zou, J.; Zhang, S.; Zhang, W.; Li, G.; Chen, Z.; Zhai, W.; Zhao, X.; Pan, X.; Xie, Q.; Zhu, L. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 2006, 48, 687–698.
- Johnson, X.; Brcich, T.; Dun, E.A.; Goussot, M.; Haurogné, K.; Beveridge, C.A.; Rameau, C. Branching Genes Are Conserved across Species. Genes Controlling a Novel Signal in Pea Are Coregulated by Other Long-Distance Signals. Plant Physiol. 2006, 142, 1014–1026.
- Foo, E.; Bullier, E.; Goussot, M.; Foucher, F.; Rameau, C.; Beveridge, C.A. The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 2005, 17, 464–474.
- Bainbridge, K.; Sorefan, K.; Ward, S.; Leyser, O. Hormonally controlled expression of the Arabidopsis MAX4 shoot branching regulatory gene. Plant J. 2005, 44, 569–580.
- Domagalska, M.A.; Leyser, O. Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 2011, 12, 211–221.
- Bennett, T.; Leyser, O. Something on the Side: Axillary Meristems and Plant Development. Plant Mol. Biol. 2006, 60, 843–854.
- Prusinkiewicz, P.; Crawford, S.; Smith, R.S.; Ljung, K.; Bennett, T.; Ongaro, V.; Leyser, O. Control of bud activation by an auxin transport switch. Proc. Natl. Acad. Sci. USA 2009, 106, 17431–17436.
- Crawford, S.; Shinohara, N.; Sieberer, T.; Williamson, L.; George, G.; Hepworth, J.; Müller, D.; Domagalska, M.A.; Leyser, O. Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 2010, 137, 2905–2913.
- Wang, Y.; Wang, J.; Shi, B.; Yu, T.; Qi, J.; Meyerowitz, E.M.; Jiao, Y. The Stem Cell Niche in Leaf Axils Is Established by Auxin and Cytokinin in Arabidopsis. Plant Cell 2014, 26, 2055–2067.
- Bangerth, F. Response of cytokinin concentration in the xylem exudate of bean (Phaseolus vulgaris L.) plants to decapitation and auxin treatment, and relationship to apical dominance. Planta 1994, 194, 439–442.
- Nordström, A.; Tarkowski, P.; Tarkowska, D.; Norbaek, R.; Åstot, C.; Dolezal, K.; Sandberg, G. Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: A factor of potential importance for auxin–cytokinin-regulated development. Proc. Natl. Acad. Sci. USA 2004, 101, 8039–8044.
- Ferguson, B.J.; Beveridge, C.A. Roles for auxin, cytokinin, and strigolactone in regulating shoot branching. Plant Physiol. 2009, 149, 1929–1944.
- Li, C.J.; Guevara, E.; Herrera, J.; Bangerth, F. Effect of apex excision and replacement by 1-naphthylacetic acid on cytokinin concentration and apical dominance in pea plants. Physiol. Plant. 1995, 94, 465–469.
- Rolland, F.; Baena-Gonzalez, E.; Sheen, J. Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 2006, 57, 675–709.
- Smeekens, S.; Ma, J.; Hanson, J.; Rolland, F. Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 2010, 13, 273–278.
- Wind, J.; Smeekens, S.; Hanson, J. Sucrose: Metabolite and signaling molecule. Phytochemistry 2010, 71, 1610–1614.
- Mason, M.G.; Ross, J.J.; Babst, B.A.; Wienclaw, B.N.; Beveridge, C.A. Sugar demand, not auxin, is the initial regulator of apical dominance. Proc. Natl. Acad. Sci. USA 2014, 111, 6092–6097.
- Barbier, F.F.; Lunn, J.E.; Beveridge, C.A. Ready, steady, go! A sugar hit starts the race to shoot branching. Curr. Opin. Plant Biol. 2015, 25, 39–45.
- Morris, S.E.; Cox, M.C.; Ross, J.J.; Krisantini, S.; Beveridge, C.A. Auxin dynamics after decapitation are not correlated with the initial growth of axillary buds. Plant Physiol. 2005, 138, 1665–1672.
- Kebrom, T.H.; Chandler, P.M.; Swain, S.M.; King, R.W.; Richards, R.A.; Spielmeyer, W. Inhibition of tiller bud outgrowth in the tin mutant of wheat is associated with precocious internode development. Plant Physiol. 2012, 160, 308–318.
- Koumoto, T.; Shimada, H.; Kusano, H.; She, K.-C.; Iwamoto, M.; Takano, M. Rice monoculm mutation moc2, which inhibits outgrowth of the second tillers, is ascribed to lack of a fructose-1, 6-bisphosphatase. Plant Biotechnol. 2013, 30, 47–56.
- Barbier, F.; Péron, T.; Lecerf, M.; Perez-Garcia, M.-D.; Barrière, Q.; Rolčík, J.; Boutet-Mercey, S.; Citerne, S.; Lemoine, R.; Porcheron, B. Sucrose is an early modulator of the key hormonal mechanisms controlling bud outgrowth in Rosa hybrida. J. Exp. Bot. 2015, 66, 2569–2582.
- Rabot, A.; Henry, C.; Ben Baaziz, K.; Mortreau, E.; Azri, W.; Lothier, J.; Hamama, L.; Boummaza, R.; Leduc, N.; Pelleschi-Travier, S.; et al. Insight into the Role of Sugars in Bud Burst Under Light in the Rose. Plant Cell Physiol. 2012, 53, 1068–1082.
- Ongaro, V.; Leyser, O. Hormonal control of shoot branching. J. Exp. Bot. 2007, 59, 67–74.
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139.
- Evers, J.B.; Vos, J.; Andrieu, B.; Struik, P.C. Cessation of Tillering in Spring Wheat in Relation to Light Interception and Red: Far-red Ratio. Ann. Bot. 2006, 97, 649–658.
- Girault, T.; Bergougnoux, V.; Combes, D.; Viemont, J.-D.; Leduc, N. Light controls shoot meristem organogenic activity and leaf primordia growth during bud burst in Rosa sp. Plant Cell Environ. 2008, 31, 1534–1544.
- Stirnberg, P.; Van De Sande, K.; Leyser, H.M.O. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 2002, 129, 1131–1141.
- Beveridge, C.A.; Weller, J.L.; Singer, S.R.; Hofer, J.M.I. Axillary Meristem Development. Budding Relationships between Networks Controlling Flowering, Branching, and Photoperiod Responsiveness. Plant Physiol. 2003, 131, 927–934.
- Kebrom, T.H.; Brutnell, T.P. The molecular analysis of the shade avoidance syndrome in the grasses has begun. J. Exp. Bot. 2007, 58, 3079–3089.
- Ballaré, C.L.; Scopel, A.L.; Sánchez, R.A. Far-red radiation reflected from adjacent leaves: An early signal of competition in plant canopies. Science 1990, 247, 329–332.
- Smith, H.; Whitelam, G.C. The shade avoidance syndrome: Multiple responses mediated by multiple phytochromes. Plant Cell Environ. 1997, 20, 840–844.
- Finlayson, S.A.; Krishnareddy, S.R.; Kebrom, T.H.; Casal, J.J. Phytochrome Regulation of Branching in Arabidopsis. Plant Physiol. 2010, 152, 1914–1927.
- Kebrom, T.H.; Burson, B.L.; Finlayson, S.A. Phytochrome B Represses Teosinte Branched1 Expression and Induces Sorghum Axillary Bud Outgrowth in Response to Light Signals. Plant Physiol. 2006, 140, 1109–1117.
- Luo, L.; Zhang, Y.; Xu, G. How does nitrogen shape plant architecture? J. Exp. Bot. 2020, 71, 4415–4427.
- Luo, Z.; Janssen, B.J.; Snowden, K.C. The molecular and genetic regulation of shoot branching. Plant Physiol. 2021, 187, 1033–1044.
- Huang, W.; Bai, G.; Wang, J.; Zhu, W.; Zeng, Q.; Lu, K.; Sun, S.; Fang, Z. Two splicing variants of OsNPF7.7 regulate shoot branching and nitrogen utilization efficiency in rice. Front. Plant Sci. 2018, 9, 300.
- Gao, Z.; Wang, Y.; Chen, G.; Zhang, A.; Yang, S.; Shang, L.; Wang, D.; Ruan, B.; Liu, C.; Jiang, H.; et al. The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency. Nat. Commun. 2019, 10, 5207.
- Gao, Y.; Qi, S.; Wang, Y. Nitrate signaling and use efficiency in crops. Plant Commun. 2022, 3, 100353.
- Wu, K.; Wang, S.; Song, W.; Zhang, J.; Wang, Y.; Liu, Q.; Yu, J.; Ye, Y.; Li, S.; Chen, J.; et al. Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science 2020, 367, eaaz2046.
- Sun, H.; Qian, Q.; Wu, K.; Luo, J.; Wang, S.; Zhang, C.; Ma, Y.; Liu, Q.; Huang, X.; Yuan, Q.; et al. Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat. Genet. 2014, 46, 652–656.
- Drummond, R.S.M.; Janssen, B.J.; Luo, Z.; Oplaat, C.; Ledger, S.E.; Wohlers, M.W.; Snowden, K.C. Environmental Control of Branching in Petunia. Plant Physiol. 2015, 168, 735–751.
- Yoneyama, K.; Xie, X.; Kim, H.I.; Kisugi, T.; Nomura, T.; Sekimoto, H.; Yokota, T.; Yoneyama, K. How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta 2012, 235, 1197–1207.
- Kamada-Nobusada, T.; Makita, N.; Kojima, M.; Sakakibara, H. Nitrogen-Dependent Regulation of De Novo Cytokinin Biosynthesis in Rice: The Role of Glutamine Metabolism as an Additional Signal. Plant Cell Physiol. 2013, 54, 1881–1893.
- Yoneyama, K.; Xie, X.; Kisugi, T.; Nomura, T.; Yoneyama, K. Nitrogen and phosphorus fertilization negatively affects strigolactone production and exudation in sorghum. Planta 2013, 238, 885–894.
- Paz-Ares, J.; Puga, M.I.; Rojas-Triana, M.; Martinez-Hevia, I.; Diaz, S.; Poza-Carrión, C.; Miñambres, M.; Leyva, A. Plant adaptation to low phosphorus availability: Core signaling, crosstalks, and applied implications. Mol. Plant 2022, 15, 104–124.
- Yuan, K.; Zhang, H.; Yu, C.; Luo, N.; Yan, J.; Zheng, S.; Hu, Q.; Zhang, D.; Kou, L.; Meng, X.; et al. Low phosphorus promotes NSP1-NSP2 heterodimerization to enhance strigolactone biosynthesis and regulate shoot and root architectures in rice. Mol. Plant 2023.
- Fioreze, S.L.; Castoldi, G.; Pivetta, L.A.; Pivetta, L.G.; Fernandes, D.M.; Büll, L.T. Tillering of two wheat genotypes as affected by phosphorus levels. Acta Sci. Agron. 2012, 34, 331–338.
- Umehara, M.; Hanada, A.; Magome, H.; Takeda-Kamiya, N.; Yamaguchi, S. Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol. 2010, 51, 1118–1126.
- Chérel, I. Regulation of K+ channel activities in plants: From physiological to molecular aspects. J. Exp. Bot. 2004, 55, 337–351.
- Yang, T.; Feng, H.; Zhang, S.; Xiao, H.; Hu, Q.; Chen, G.; Xuan, W.; Moran, N.; Murphy, A.; Yu, L.; et al. The Potassium Transporter OsHAK5 Alters Rice Architecture via ATP-Dependent Transmembrane Auxin Fluxes. Plant Commun. 2020, 1, 100052.
- Xu, Y.; Zhang, S.; Guo, H.; Wang, S.; Xu, L.; Li, C.; Qian, Q.; Chen, F.; Geisler, M.; Qi, Y.; et al. OsABCB14 functions in auxin transport and iron homeostasis in rice (Oryza sativa L.). Plant J. Cell Mol. Biol. 2014, 79, 106–117.
- Panda, D.; Mishra, S.S.; Behera, P.K. Drought tolerance in rice: Focus on recent mechanisms and approaches. Rice Sci. 2021, 28, 119–132.
- Sunkar, R.; Zhu, J.-K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019.
- Prasanth, V.V.; Babu, M.S.; Basava, R.K.; Tripura Venkata, V.G.N.; Mangrauthia, S.K.; Voleti, S.R.; Neelamraju, S. Trait and Marker Associations in Oryza nivara and O. rufipogon Derived Rice Lines under Two Different Heat Stress Conditions. Front. Plant Sci. 2017, 8, 1819.
- Harsant, J.; Pavlovic, L.; Chiu, G.; Sultmanis, S.; Sage, T.L. High temperature stress and its effect on pollen development and morphological components of harvest index in the C3 model grass Brachypodium distachyon. J. Exp. Bot. 2013, 64, 2971–2983.
- Wang, Y.; Sun, F.; Cao, H.; Peng, H.; Ni, Z.; Sun, Q.; Yao, Y. TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS ONE 2012, 7, e48445.
- Chen, L.; Zhao, Y.; Xu, S.; Zhang, Z.; Xu, Y.; Zhang, J.; Chong, K. OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol. 2018, 218, 219–231.
More
