You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 1 by Rashid Minhas and Version 2 by Sirius Huang.
The heart is composed of muscle cells called cardiomyocytes, including a specialized population named pacemaker cells that form the cardiac conduction system (CCS), which is responsible for generating the action potential dictating heart contractions. Failure of the CCS system leads to cardiac arrhythmias, which require complicated therapies and often the surgical implantation of electrical pacemakers.
- cardiac conduction system
- pacemaker
- gene regulatory network
1. Contracting Cardiac Muscle
The heart, a muscular organ, orchestrates blood circulation throughout the body by generating and coordinating electrical impulses. These impulses are controlled by the cardiac conduction system (CCS), which consists of myogenic components that regulate the contractions of the atria and ventricles [1][2][3][4][1,2,3,4]. In higher vertebrates, the CCS is divided into specific regions: the slow-conducting structures, such as the sinoatrial node (SAN); the primary site for pacemaker cardiomyocytes (CMs), often referred to as the “natural pacemakers” of the heart, which are located between the superior vena cava and the right atrium; the secondary pacemaker, the atrioventricular node (AVN), which is situated within the atrioventricular septum; and the fast-conducting ventricular conducting system (VCS), which includes the atrioventricular bundle (AVB, also known as the His-Bundle), the right and left bundle branches (BBs), and the Purkinje fiber network (PFN) (which is responsible for ventricular coordination) [3][5][6][3,5,6]. Pacemaker cells within the CCS are a specialized population with a unique and vital role. They have the remarkable capability to spontaneously generate regular electrical impulses, effectively setting the pace for the entire heart. These rhythmic depolarization and repolarization cycles in pacemaker cells are responsible for initiating each heartbeat and maintaining its regularity [4]. These electrical signals rapidly travel through the atrial cardiomyocytes, thus initiating atrial contraction [7]. Subsequently, they navigate through slower-conducting tissues within the AVN, introducing a deliberate delay before transmitting to the His-Purkinje fibers, thereby effectively coordinating ventricular contraction [4][8][4,8] (Figure 1).
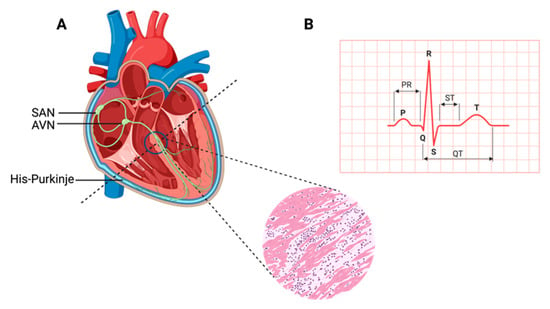
Figure 1. Cardiac contraction and the histology of cardiac muscle. (A) Schematic representation of the components of the cardiac conduction system (CCS) in a human heart. In the inset picture, a cross-section of the human heart muscle is shown with binucleated cardiomyocytes. The various components of the CCS (in green) are labeled: the sinoatrial node (SAN), found at the junction of the superior caval vein and right atrium, generates the impulse that then travels to the atrioventricular node (AVN). Propagation occurs through the left and right bundle branches of His-Purkinje, leading to ventricular contraction. (B) An electrocardiogram representing the recording of the electrical activity of the heart. The upper chambers of the heart (atria) begin to beat when the first wave of the ECG, labeled P, appears. The lower chambers of the heart (ventricles) are represented by the QRS complex as an electrical current flow. The electrical current spreads back over the ventricles in the opposite direction during the recovery phase, which is represented by the T wave.
Cardiac muscle development is a complex and tightly regulated process involving the differentiation and specialization of various cell types, including CMs and pacemaker cells. Notably, diverse vertebrate species exhibit a remarkable spectrum of cardiac development timelines. In a developing human embryo, the initial indications of heart muscle contractions typically emerge around embryonic day 22, which marks the third week of gestation. This coincides with the evolution of the first heart field into the heart tube [9]. In comparison, mice exhibit an earlier onset of heart muscle contractions, typically occurring around embryonic days E8 to E9 [10][11][10,11]. However, zebrafish exhibit a unique timeline, possessing distinct heart muscle anatomy from humans, mice, or chickens. Contraction in zebrafish begins as early as 22 h after fertilization. In chickens, contraction initiates at approximately HH10 to HH11 [12][13][14][12,13,14], and each species reflects the intricacies of its cardiac development.
Studies from mammalian, avian, and fish model systems have shown that each CCS component consists of a specialized group of CMs with distinctive morphological and electrophysiological properties, as well as transcriptional profiles [15]. This specialized development of the CCS and pacemaker cells plays a pivotal role in establishing a fully functional heart. However, disruptions or defects in this developmental process can lead to cardiac arrhythmias, including conditions like Brugada syndrome, long QT syndrome, and sudden cardiac death [16][17][16,17]. These arrhythmias are characterized by irregular or slow heartbeats, thus ultimately compromising the heart’s ability to efficiently pump blood. Importantly, some of these arrhythmias can further complicate the treatment of congenital cardiac conditions, which often require therapeutics like ion channel blockers, or surgical interventions such as ablation or electronic pacemaker implantation [4]. However, the limitations associated with conventional pacemakers have highlighted the pressing need for alternative pacemaker solutions, leading to the emergence of biological pacemakers as a promising avenue for improving the management of cardiac arrhythmias.
2. Key Transcription Factors Involved in CCS Development
Exploring CCS development and homeostasis is reliant on transcriptional and regulatory networks that are embryonic-stage-dependent, dose-dependent, and tissue-dependent [18][19][20][34,35,36]. A cascade of transcription factors, SHOX2, BMP4, NKX2-5, ID2, ISL1, GATA4, HAND1, IRX3, and various T-box transcriptions factors are instrumental to the divergence in myocyte development. Below are sub-sections that show the evidence as to why these transcription factors are important.
2.1. Short Stature Homeobox 2 (SHOX2) and Bone Morphogenic Protein 4 (BMP4)
SHOX2 and BMP4 are recognized as pivotal factors contributing to the formation of the SAN, a critical element in heart rhythm regulation [4][21][4,37]. Dysfunction of the SAN can precipitate various cardiac arrhythmias, including bradycardic arrhythmias [22][38]. The SHOX2 transcription factor holds essential significance in both SAN development and differentiation. Its wide expression throughout the body, including the heart muscle, underscores its multifaceted role [22][38]. Conversely, BMPs, a subgroup of signaling molecules within the Transforming Growth Factor β superfamily, play a vital role in pacemaker development. BMP4, in particular, assumes a central position in embryonic heart development by promoting fibroblast reprogramming into cardiomyocytes with pacemaker activity [23][39]. Notably, SHOX2 exerts influence over BMP4, where their expressions overlap [24][40]. Among the remarkable functions of BMP4 are that it takes a lead role in driving the differentiation of cardiac pacemaker cells [25][41]. Illumination from epistatic genetic experiments conducted in Xenopus has unveiled a direct interaction between SHOX2 and the BMP4 promoter. The closely coordinated expression patterns of BMP4 and SHOX2 are especially conspicuous in the SAN during embryonic development [24][40].
2.2. T-Box Transcription Factor 5 (TBX5), NK2 Homeobox 5 (NKX2-5), and Inhibitor of DNA Binding 2 (ID2)
TBX5, NKX2-5, and ID2 play indispensable roles in the development of the atrioventricular bundle and bundle branches [4][26][4,42]. The transcription factor TBX5, while having diverse functions across the body, holds a pivotal role in cardiac development [26][42]. Mutations in TBX5 have been associated with cardiac defects in the septa and CCS. During early embryonic cardiac development, TBX5 functions as a transcriptional activator for genes involved in cardiomyocyte maturation [26][42]. In later cardiac development stages, TBX5 shifts its focus to the structure of the CCS and the maintenance of cardiomyocyte maturation [26][42]. NKX2-5, a cardiac homeobox transcription factor with an expression spanning the cardiac system, plays a crucial role in regulating cardiac development and function [27][43]. Mutations in NKX2-5 result in cardiac defects and atrioventricular conduction irregularities. Throughout cardiac development, NKX2-5 is instrumental in regulating the function of working and in conducting myocytes within the atria, often in coordination with the Notch signaling pathway [27][43]. ID2, another cardiac transcription factor, is initially detected in areas like the neural crest, in inflow and outflow tracts, and in neurons around the aorta and pulmonary artery [28][29][44,45]. In later developmental stages, ID2 expression becomes apparent in the atrioventricular bundle around E12.5 and subsequently in the bundle branches by, approximately, E16.5 [26][29][42,45].
2.3. T-Box Transcription Factor 3 (TBX3)
TBX3, a vital player within the CCS, is integral for repressing atrial differentiation and maintaining proper cardiac function [30][46]. Various studies have linked noncoding variants near TBX3 expression to alterations in PR interval and QRS duration, underscoring its impact on atrioventricular conduction [31][32][33][34][35][36][47,48,49,50,51,52]. TBX3 is primarily found in the SAN, which is a part of the heart’s electrical system. It plays a major role in regulating the genes active in the SAN while actively suppressing genes associated with atrial function, ensuring that the SAN retains its pacemaker function and does not become atrial tissue [4]. When TBX3 is introduced where it is not typically found, it prompts the development of functional pacemaker cells within the atria [37][38][53,54]. In essence, TBX3 transforms regular cardiac cells into pacemaker-like cells within its domain of influence.
In the nearby developing atrial heart tissue, Nkx2-5 has an opposing role. Nkx2-5 represses the expression of TBX3 and another gene called Hcn4. This is consistent with observations in embryos lacking Nkx2-5, which show abnormal expressions of TBX3 and Hcn4 in the heart tube [39][55]. Conversely, introducing extra Nkx2-5 into heart muscle cells, including those in the SAN, prevents the proper formation of the SAN [40][56]. This indicates that Nkx2-5 acts to confine the influence of TBX3 and Hcn4 to specific areas of the heart.
Interestingly, the absence of Nkx2-5 in the SAN, while present in other heart muscle cells, provides a valuable tool for identifying SAN cells in laboratory-grown human ESCs. When scientists coax human ESCs into becoming heart cells, they produce both cells similar to those found in the heart’s chambers (NKX2-5+) and pacemaker-like cells that lack NKX2-5 expression (NKX2-5−) [41][57]. This research enhances our understanding of cardiac development and the roles of these critical transcription factors.
2.4. T-box Transcription Factor 18 (TBX18)
TBX18 plays a pivotal role in heart muscle development, particularly in shaping the structure and formation of the SAN [25][42][41,58]. Its expression is essential for early SAN specification, and it generates pacemaker activity during the initial phases of embryonic heart muscle formation [30][43][44][46,59,60]. Surprisingly, when Tbx18 is deficient in mice (which leads to underdeveloped sinus venosus and SAN structures), these mice do not display significant bradycardia (slow heart rate). Intriguingly, even in the presence of this deficiency, the SAN gene program remains intact in their underdeveloped SANs. This suggests that while Tbx18 may not directly control the SAN gene program, it plays an essential role in ensuring the proper formation and deployment of progenitor cells [30][46]. However, when Tbx18 is artificially introduced into ventricular myocytes via a viral method, it has a distinct impact. Specifically, it reduces the expression of connexin 43 (Cx43), a protein responsible for gap junction intercellular communication between cells, which regulates cell death, proliferation, and differentiation (while not affecting Cx40 and Cx45), in these ventricular myocytes [45][61]. In the ventricles of pigs and guinea pigs, the introduction of Tbx18 leads to a phenomenon called ‘reprogramming,’ where ventricular myocytes start to exhibit pacemaker-like properties and generate ectopic pacemaker activity. Alongside this reprogramming, there is a suppression of Cx43 and natriuretic peptide A (Nppa), as well as an increase in Hcn4 expression [46][47][62,63]. It is speculated that the differences observed between the loss and gain of Tbx18 function experiments can be attributed to the fact that Tbx18, which primarily acts as a repressor T-box factor [48][64], mimics the function of Tbx3 when overexpressed [4].
2.5. ISLET-1 (ISL1)
ISL1, a transcription factor, fulfills diverse roles across multiple organs during embryonic development, and, within cardiac development, it serves as a marker for second heart field progenitors [49][65]. Its expression is detectable as early as E7 in mouse heart development, and its pattern shifts as development progresses, with the expression being observed in the SAN from postnatal stages through to adulthood [50][66]. In zebrafish, Isl1 is a marker for pacemaker cells located at the junction of the sinus venosus and atrium, where it is necessary for normal pacemaker function and development [51][67]. In mice, Isl1 is indispensable for the proliferation and proper functioning of SAN cells, and its specific deletion within the SAN results in embryonic lethality [21][37]. Notably, the absence of Isl1 in mice leads to the downregulation of the key regulators involved in SAN development, such as TBX3, SHOX2, and BMP4, as well as ion channels that are crucial for SAN function, including HCN4, HCN1, and Cacna1g [21][52][37,68]. Conversely, when Isl1 is overexpressed in the cardiomyocytes derived from ESCs, it upregulates the genes associated with the SAN while downregulating genes linked to chamber myocardia [53][69]. Remarkably, Isl1 is a target of SHOX2 within the SAN, and it can rescue the bradycardia phenotype that results from SHOX2 deficiency [54][70]. This emphasizes the pivotal role of ISL1 in the development and regulation of pacemaker cells within the heart’s SAN.
2.6. GATA4
GATA4 functions as a crucial regulator of cardiomyocyte proliferation and differentiation. It exhibits high expression levels until birth and remains detectable in all cardiomyocytes [55][71]. It persists in its expression until approximately one week after birth, remaining easily detectable in cardiomyocytes and other cardiac cells, including those within the outflow tract (OFT), septa, and valves [55][71]. However, the absence of GATA4 in mice leads to embryonic lethality at E8.5, which is accompanied by cardia bifida, underscoring its crucial function in early heart formation [55][56][71,72]. Different studies have employed conditional knockout models. These models have shed light on GATA4’s dosage-sensitive role. Introducing LoxP sites to the GATA4 gene results in decreased expressions of around 20%, leading to structural heart defects [57][73]. Deleting GATA4 from cardiomyocytes using the Nkx2.5-Cre driver, occurring around E9.5, leads to myocardial thinning, the absence of mesenchymal cells in the endocardial cushions, a hypoplastic right ventricle, and embryonic lethality by E11.5 [58][74]. In contrast, the removal of Gata4 from endocardial cells through the Tie2-Cre driver results in embryonic lethality at E12.5 due to impaired epithelial-to-mesenchymal transformation (EMT), thus contributing to underdeveloped atrioventricular cushions [59][75]. Although GATA4 deletion when using the βMHC-Cre driver at E17.5 results in viable and fertile mice, it makes them susceptible to left ventricular dysfunction and dilation [60][76]. These findings underscore GATA4’s multifaceted role in regulating various aspects of cardiac development, including pacemaker cell development, as well as emphasize its importance in maintaining proper cardiac function.
On the other hand, GATA6 plays a pivotal role in the development of the SAN. Mutations in the GATA6 gene can lead to dysfunction in SAN patterning and size, ultimately contributing to the occurrence of arrhythmias [61][77]. This includes reduced levels of essential regulators for pacemaker cells like TBX3 and TBX5, which are accompanied by an increase in genes associated with the atria, such as Nkx2.5 and Nppa. Additionally, the arrangement of the SAN seems disturbed, particularly in the loss of HCN4+ pacemaker cells, which are mainly present in the head region [61][77]. While GATA6’s involvement in pacemaker cell differentiation is clear, it appears to have different functions depending on the type of cells within the SAN. In ISL1+ myocytes and HCN4+ conduction cells, GATA6 acts as an activator for the genetic program needed for pacemaker cell differentiation. It functions upstream of various transcriptional regulators in the SAN, including TBX3, TBX5, and TBX18 [61][77]. On the contrary, in endothelial cells, GATA6 probably regulates pacemaker cell differentiation indirectly, potentially through the influence on paracrine factors like EDN1, which plays a role in SAN cell differentiation [62][78]. An analysis of transcripts showed reduced levels of both EDN1 and one of its receptors, EDNRB, in the hearts of Gata6+/− mice as early as E11.5. This suggests that one way in which GATA6 contributes to the regulation of pacemaker cell differentiation in these cells is by modulating the activity of EDN1 [61][77].
2.7. HAND1
HAND1 plays a critical role in the specification and differentiation of embryonic structures, including the cardiac muscle of the heart [63][79]. It functions as an essential regulator for determining the fate of cardiac precursor cells, and it is involved in morphogenesis—a process controlled by the BMP signaling pathway [64][80]. Mutations in the HAND1 gene have been linked to congenital heart disease, highlighting its significance in heart development [65][66][81,82]. Moreover, recent research has revealed that BMP signaling can activate HAND1 regulation, further illuminating its role in heart muscle development [63][79].
2.8. IRX3
IRX3 plays a crucial role in regulating rapid electrical propagation within the ventricular conduction system by facilitating the transcription of Cx40 and Cx43 genes [67][83]. The development of the ventricular conduction system is tightly controlled by the activation of various transcription factors, including NKX2-5, TBX3, TBX5, and ID2 [26][67][68][69][70][71][72][42,83,84,85,86,87,88]. The dysregulation or loss of these transcription factors can result in a range of cardiac defects, particularly NKX2-5 and TBX5 loss, which can elevate the risk of arrhythmias [67][83]. IRX5 exhibits a gradient of expression within the ventricular myocardium, with the epicardium showing lower expression and the endocardium displaying higher expression levels [73][89]. Mutations in IRX5 are associated with an increased susceptibility to arrhythmias due to abnormal repolarization in the ventricular conduction system, which is influenced by the absence of a homeostatic Kv4.2 gradient [73][74][89,90].
The identification and understanding of the transcription factors involved in cardiac development, particularly in the context of the CCS, play a pivotal role in bridging the gap between existing knowledge and the practical applications in reprogramming strategies. Understanding the transcription factors’ roles in cardiac development provides a roadmap for designing targeted reprogramming approaches. By harnessing these insights, the efficiency and effectiveness of reprogramming strategies can be optimized, bringing us closer to the practical application of biological pacemakers and other therapeutic interventions for cardiac conduction disorders.
