You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Fanny Huang and Version 1 by Yongmin Cho.
Single-molecule imaging technologies, especially those based on fluorescence, have been developed to probe both the equilibrium and dynamic properties of biomolecules at the single-molecular and quantitative levels. Lab-on-a-chip techniques have emerged as valuable tools in single-molecule fluorescence imaging, offering numerous advantages and enabling new possibilities for experimental design and analysis.
- single-molecule fluorescence imaging
- lab-on-a-chip techniques
1. Introduction
Imaging techniques provide powerful tools to visualize and quantify molecular interactions, cellular dynamics, and tissue architecture and are therefore instrumental in advancing our understanding of biological systems [1,2,3,4,5,6,7,8,9,10,11,12][1][2][3][4][5][6][7][8][9][10][11][12]. Certain imaging techniques can directly observe individual biomolecules such as oligonucleotides, proteins, and protein complexes. These single-molecule imaging techniques can provide information on the heterogeneity of the system which can often be difficult to determine using other methods. In recent years, single-molecule imaging with total internal reflection fluorescence (TIRF) has gained significant popularity due to its accessibility and high sensitivity in probing the properties of biomolecules. By enabling the visualization and tracking of individual molecules in exceptional spatial and temporal resolutions, TIRF-based single-molecule imaging has opened up new avenues for studying complex biological processes, including protein folding, protein–protein interactions, DNA replication, and cellular signaling [13,14,15,16,17,18,19,20,21,22][13][14][15][16][17][18][19][20][21][22].
2. Optical Systems for Single-Molecule Fluorescence Imaging
Fluorescence single-molecule imaging techniques rely on the utilization of optical radiation to probe individual molecules within a liquid or solid sample. To achieve successful single-molecule imaging, two key requirements must be met: (1) ensuring that resonant molecules are spatially resolved by the detector, and (2) providing a sufficient signal-to-noise ratio (SNR) for the single-molecule signal within a reasonable averaging time [22]. Consequently, a fundamental prerequisite for conducting single-molecule observations is to dilute the concentration of the target molecule of interest to exceedingly low levels (typically < 100 nM). The detection of single molecules via fluorescence-based methods demands careful optimization of the signal-to-noise ratio. Maximizing the signal requires the selection of an impurity molecule with the highest possible fluorescence quantum efficiency. This approach harnesses recent advancements in fluorescence imaging techniques, including TIRF microscopy [23,24[23][24][25],25], super-resolution microscopy, and single-molecule localization microscopy [26,27,28,29,30,31,32][26][27][28][29][30][31][32]. TIRF microscopy, one of the most commonly employed tools in single-molecule fluorescence microscopy, capitalizes on the principle of total internal reflection. This phenomenon occurs when a laser beam strikes the interface between a medium with a higher refractive index (typically glass) and a medium with a lower refractive index (such as a sample solution) at an angle greater than the critical angle (Figure 1A). As a result, an evanescent wave is generated, which excites fluorophores in the immediate vicinity of the interface, facilitating the visualization of single molecules near the sample surface. TIRF microscopy is practically implemented by using either a quartz prism or the microscope objective to generate the evanescent field and illuminate surface-immobilized molecules (Figure 1B). TIRF microscopy offers exceptional optical sectioning and background suppression, leading to a high signal-to-noise ratio.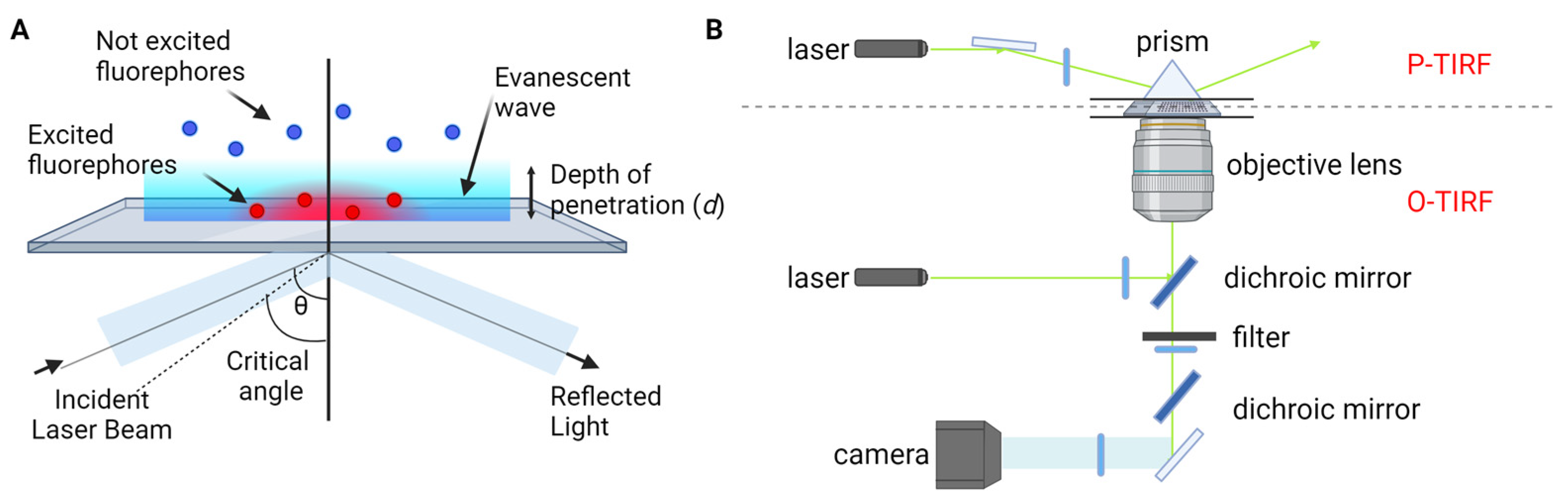
Figure 1. TIRF microscopy for single-molecule fluorescence imaging. (A) Principle of TIRF microscopy. (B) Types of TIRF microscopy: prism-type (P-TIRF) or objective-type (O-TIRF).
3. Application of Lab-on-a-Chip Techniques for Single-Molecule Fluorescence Imaging
Lab-on-a-chip techniques have emerged as valuable tools in single-molecule fluorescence imaging, offering numerous advantages and enabling new possibilities for experimental design and analysis [87,88,89,90,91,92,93][34][35][36][37][38][39][40]. Lab-on-a-chip devices provide unparalleled control and manipulation capabilities, enabling precise management of fluid flow and sample handling in single-molecule imaging experiments through the utilization of a laminar flow regime. The controlled flow within microfluidic channels not only facilitates efficient sample processing but also aids in reducing background noise by removing unbound or non-specifically bound molecules, thereby enhancing the SNR. Moreover, lab-on-a-chip platforms leverage miniaturized channels and chambers to create controlled microenvironments for the delivery, mixing, and incubation of samples, as well as the manipulation of individual molecules.
3.1. Enhance Signal-to-Noise Ratio
One advantage of single-molecule imaging over ensemble studies is its superior time domain resolution for investigating molecular dynamics. However, this advantage can be compromised by the limited photostability of singlet exciton emission, which is prone to bleaching and blinking due to factors like O2 and intersystem crossing. The stochastic fluctuations resulting from blinking are unrelated to the underlying biological behavior. To overcome these challenges, microfluidics has been employed by incorporating oxygen scavengers and triplet quenchers into the imaging buffer [95,96][41][42]. By carefully designing the setup, this approach has recently facilitated the shortest observation durations [97][43]. It effectively addresses the limitation of time domain resolution posed by fluidic speed, particularly during fluidic mixing. A different method was shown [98][44], wherein the imaging channels were integrated with those consistently supplied with nitrogen ventilation. Furthermore, sophisticated microfluidic architectures can reduce flow velocities immediately after mixing, enabling longer optical interrogations [99][45].
3.2. Increase Sample Concentration
As mentioned, single-molecule fluorescence imaging techniques are limited to using pico- to nanomolar concentrations to ensure that only single molecules are resonant within the laser-probed volume and provide a sufficient SNR. However, many biologically relevant processes occur at micromolar level concentrations, necessitating a reduction in the conventional observation volume by three orders of magnitude. Here, arrays of zero-mode waveguides (ZMWs) consisting of subwavelength holes in a metal film provide a means to increase sample concentrations to the micromolar range while confining the observation volume to zeptoliter dimensions [100,101,102][46][47][48]. This breakthrough enables studies in the physiological concentration range and has been successfully applied in real-time, protein–protein interactions [102][48]. ZMWs have also been utilized to investigate ribosome-mediated translation processes, allowing the observation of tRNA transit in real-time at physiological concentrations [103][49]. Additionally, ZMWs have demonstrated versatility in studying biomolecular interactions, protein receptor diffusion, and oligomerization on living cell membranes [94][50].
3.3. On-Chip Single-Molecule Manipulation
On-chip devices have been developed with the capability to spatially modulate individual molecules with nanometer or even sub-nanometer sensitivities. A notable example is the microfluidic-based “DNA curtain”, which has recently emerged as an elegant on-chip tool for investigating DNA–protein interactions [16,17,18,47,104][16][17][18][51][52]. This technique involves driving DNA molecules that are tethered to a fluidic lipid bilayer on the surface. These molecules drift downstream under the influence of flow until they encounter a thin layer of metal, which serves as a diffusion barrier. Consequently, the DNA molecules align with each other, forming what is referred to as a DNA curtain [47][51]. This fluidic-chip setup has proven highly successful in unraveling the searching modes of a DNA repair complex at DNA damage and elucidating the disruption of a transcription complex by a DNA translocase at the single-molecule level [18]. In addition, Alwan et al. utilized a microfluidics-based single-molecule live cell fluorescence imaging to study the spatiotemporal dynamics of selectin ligands on the membrane tethers and slings during cell rolling [105][53].
3.4. Microenvironment Control
Lab-on-a-chip technology also affords precise control over the microenvironment surrounding single molecules. Variables such as temperature, pH, and chemical gradients can be precisely manipulated within microfluidic devices, providing valuable insights into the impact of different conditions on the behavior and functionality of biomolecules. This level of control allows for the investigation of dynamic processes under various physiological or pathological conditions, mimicking complex biological environments. For example, Zhang et al. studied the in situ conformational response of single biomolecules such as DNA to a change in environmental solution conditions [106][54]. This level of control allows researchers to probe biomolecular interactions, enzymatic activities, and other dynamic processes with exceptional temporal resolution.
Moreover, lab-on-a-chip devices possess the remarkable capability of automation when integrated with other techniques. This integration not only minimizes experimental bias but also facilitates high-throughput screening, data acquisition, and analysis, which are indispensable for conducting large-scale single-molecule studies. By automating microfluidic processes, researchers can streamline their experiments, achieve consistent and reliable results, and analyze vast amounts of data efficiently.
References
- Lee, K.; Kim, K.; Jung, J.; Heo, J.; Cho, S.; Lee, S.; Chang, G.; Jo, Y.; Park, H.; Park, Y.; et al. Quantitative phase imaging techniques for the study of cell pathophysiology: From principles to applications. Sensors 2013, 13, 4170–4191.
- Royer, L.A.; Lemon, W.C.; Chhetri, R.K.; Wan, Y.; Coleman, M.; Myers, E.W.; Keller, P.J. Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms. Nat. Biotechnol. 2016, 34, 1267–1278.
- Wassie, A.T.; Zhao, Y.; Boyden, E.S. Expansion microscopy: Principles and uses in biological research. Nat. Methods 2019, 16, 33–41.
- Becker, W. Fluorescence lifetime imaging–techniques and applications. J. Microsc. 2012, 247, 119–136.
- Fu, D.; Zhou, J.; Zhu, W.S.; Manley, P.W.; Wang, Y.K.; Hood, T.; Wylie, A.; Xie, X.S. Imaging the intracellular distribution of tyrosine kinase inhibitors in living cells with quantitative hyperspectral stimulated Raman scattering. Nat. Chem. 2014, 6, 614–622.
- Lu, F.K.; Basu, S.; Igras, V.; Hoang, M.P.; Ji, M.; Fu, D.; Holtom, G.R.; Neel, V.A.; Freudiger, C.W.; Fisher, D.E.; et al. Label-free DNA imaging in vivo with stimulated Raman scattering microscopy. Proc. Natl. Acad. Sci. USA 2015, 112, 11624–11629.
- Roeffaers, M.B.; Zhang, X.; Freudiger, C.W.; Saar, B.G.; van Ruijven, M.; van Dalen, G.; Xiao, C.; Xie, X.S. Label-free imaging of biomolecules in food products using stimulated Raman microscopy. J. Biomed. Opt. 2010, 15, 066016.
- Shi, L.; Zheng, C.; Shen, Y.; Chen, Z.; Silveira, E.S.; Zhang, L.; Wei, M.; Liu, C.; de Sena-Tomas, C.; Targoff, K.; et al. Optical imaging of metabolic dynamics in animals. Nat. Commun. 2018, 9, 2995.
- Jalili, N.; Laxminarayana, K. A review of atomic force microscopy imaging systems: Application to molecular metrology and biological sciences. Mechatronics 2004, 14, 907–945.
- Kherlopian, A.R.; Song, T.; Duan, Q.; Neimark, M.A.; Po, M.J.; Gohagan, J.K.; Laine, A.F. A review of imaging techniques for systems biology. BMC Syst. Biol. 2008, 2, 74.
- Shashkova, S.; Leake, M.C. Single-molecule fluorescence microscopy review: Shedding new light on old problems. Biosci. Rep. 2017, 37, BSR20170031.
- Cho, Y.; Zhao, C.L.; Lu, H. Trends in high-throughput and functional neuroimaging in Caenorhabditis elegans. Wiley Interdiscip. Rev. Syst. Biol. Med. 2017, 9, e1376.
- Lu, Y.; Lee, B.-h.; King, R.W.; Finley, D.; Kirschner, M.W. Substrate degradation by the proteasome: A single-molecule kinetic analysis. Science 2015, 348, 1250834.
- Lu, Y.; Wang, W.; Kirschner, M.W. Specificity of the anaphase-promoting complex: A single-molecule study. Science 2015, 348, 1248737.
- Rief, M.; Žoldák, G. Single-molecule mechanical studies of chaperones and their clients. Biophys. Rev. 2022, 3, 041301.
- Georgescu, R.E.; Yao, N.Y.; O’Donnell, M. Single-molecule analysis of the Escherichia coli replisome and use of clamps to bypass replication barriers. FEBS Lett. 2010, 584, 2596–2605.
- Visnapuu, M.-L.; Greene, E.C. Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition. Nat. Struct. Mol. Biol. 2009, 16, 1056–1062.
- Gorman, J.; Wang, F.; Redding, S.; Plys, A.J.; Fazio, T.; Wind, S.; Alani, E.E.; Greene, E.C. Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proc. Natl. Acad. Sci. USA 2012, 109, E3074–E3083.
- Hon, J.; Lu, Y. Single-molecule methods for measuring ubiquitination and protein stability. Methods Enzymol. 2019, 619, 225–247.
- van Ginkel, J.; Filius, M.; Szczepaniak, M.; Tulinski, P.; Meyer, A.S.; Joo, C. Single-molecule peptide fingerprinting. Proc. Natl. Acad. Sci. USA 2018, 115, 3338–3343.
- Gust, A.; Zander, A.; Gietl, A.; Holzmeister, P.; Schulz, S.; Lalkens, B.; Tinnefeld, P.; Grohmann, D. A starting point for fluorescence-based single-molecule measurements in biomolecular research. Molecules 2014, 19, 15824–15865.
- Moerner, W.; Fromm, D.P. Methods of single-molecule fluorescence spectroscopy and microscopy. Rev. Sci. Instrum. 2003, 74, 3597–3619.
- Axelrod, D. Total internal reflection fluorescence microscopy. Methods Cell Biol. 1989, 30, 245–270.
- Axelrod, D. Total internal reflection fluorescence microscopy in cell biology. Traffic 2001, 2, 764–774.
- Schneckenburger, H. Total internal reflection fluorescence microscopy: Technical innovations and novel applications. Curr. Opin. Biotechnol. 2005, 16, 13–18.
- Endesfelder, U.; Heilemann, M. Art and artifacts in single-molecule localization microscopy: Beyond attractive images. Nat. Methods 2014, 11, 235–238.
- Mortensen, K.I.; Churchman, L.S.; Spudich, J.A.; Flyvbjerg, H. Optimized localization analysis for single-molecule tracking and super-resolution microscopy. Nat. Methods 2010, 7, 377–381.
- Wu, Y.-L.; Tschanz, A.; Krupnik, L.; Ries, J. Quantitative data analysis in single-molecule localization microscopy. Trends Cell Biol. 2020, 30, 837–851.
- Sage, D.; Kirshner, H.; Pengo, T.; Stuurman, N.; Min, J.; Manley, S.; Unser, M. Quantitative evaluation of software packages for single-molecule localization microscopy. Nat. Methods 2015, 12, 717–724.
- Khater, I.M.; Nabi, I.R.; Hamarneh, G. A review of super-resolution single-molecule localization microscopy cluster analysis and quantification methods. Patterns 2020, 1, 100038.
- Lelek, M.; Gyparaki, M.T.; Beliu, G.; Schueder, F.; Griffié, J.; Manley, S.; Jungmann, R.; Sauer, M.; Lakadamyali, M.; Zimmer, C. Single-molecule localization microscopy. Nat. Rev. Methods Primers 2021, 1, 39.
- Nicovich, P.R.; Owen, D.M.; Gaus, K. Turning single-molecule localization microscopy into a quantitative bioanalytical tool. Nat. Protoc. 2017, 12, 453–460.
- Roy, R.; Hohng, S.; Ha, T. A practical guide to single-molecule FRET. Nat. Methods 2008, 5, 507–516.
- Bhagat, A.A.; Bow, H.; Hou, H.W.; Tan, S.J.; Han, J.; Lim, C.T. Microfluidics for cell separation. Med. Biol. Eng. Comput. 2010, 48, 999–1014.
- Kuntaegowdanahalli, S.S.; Bhagat, A.A.; Kumar, G.; Papautsky, I. Inertial microfluidics for continuous particle separation in spiral microchannels. Lab. Chip 2009, 9, 2973–2980.
- Lu, Y.; Yang, L.; Wei, W.; Shi, Q. Microchip-based single-cell functional proteomics for biomedical applications. Lab. Chip 2017, 17, 1250–1263.
- Shields, C.W.; Reyes, C.D.; Lopez, G.P. Microfluidic cell sorting: A review of the advances in the separation of cells from debulking to rare cell isolation. Lab. Chip 2015, 15, 1230–1249.
- Xi, H.D.; Zheng, H.; Guo, W.; Ganan-Calvo, A.M.; Ai, Y.; Tsao, C.W.; Zhou, J.; Li, W.; Huang, Y.; Nguyen, N.T.; et al. Active droplet sorting in microfluidics: A review. Lab. Chip 2017, 17, 751–771.
- Yin, H.; Marshall, D. Microfluidics for single cell analysis. Curr. Opin. Biotechnol. 2012, 23, 110–119.
- Young, E.W.; Beebe, D.J. Fundamentals of microfluidic cell culture in controlled microenvironments. Chem. Soc. Rev. 2010, 39, 1036–1048.
- Rasnik, I.; McKinney, S.A.; Ha, T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat. Methods 2006, 3, 891–893.
- Vasdekis, A.E.; Laporte, G.P. Enhancing single molecule imaging in optofluidics and microfluidics. Int. J. Mol. Sci. 2011, 12, 5135–5156.
- Campos, L.A.; Liu, J.; Wang, X.; Ramanathan, R.; English, D.S.; Munoz, V. A photoprotection strategy for microsecond-resolution single-molecule fluorescence spectroscopy. Nat. Methods 2011, 8, 143–146.
- Lemke, E.A.; Gambin, Y.; Vandelinder, V.; Brustad, E.M.; Liu, H.-W.; Schultz, P.G.; Groisman, A.; Deniz, A.A. Microfluidic device for single-molecule experiments with enhanced photostability. J. Am. Chem. Soc. 2009, 131, 13610–13612.
- Gambin, Y.; VanDelinder, V.; Ferreon, A.C.; Lemke, E.A.; Groisman, A.; Deniz, A.A. Visualizing a one-way protein encounter complex by ultrafast single-molecule mixing. Nat. Methods 2011, 8, 239–241.
- Levene, M.J.; Korlach, J.; Turner, S.W.; Foquet, M.; Craighead, H.G.; Webb, W.W. Zero-mode waveguides for single-molecule analysis at high concentrations. Science 2003, 299, 682–686.
- Zhu, P.; Craighead, H.G. Zero-mode waveguides for single-molecule analysis. Annu. Rev. Biophys. 2012, 41, 269–293.
- Miyake, T.; Tanii, T.; Sonobe, H.; Akahori, R.; Shimamoto, N.; Ueno, T.; Funatsu, T.; Ohdomari, I. Real-time imaging of single-molecule fluorescence with a zero-mode waveguide for the analysis of protein− protein interaction. Anal. Chem. 2008, 80, 6018–6022.
- Uemura, S.; Aitken, C.E.; Korlach, J.; Flusberg, B.A.; Turner, S.W.; Puglisi, J.D. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature 2010, 464, 1012–1017.
- Zhao, Y.; Chen, D.; Yue, H.; French, J.B.; Rufo, J.; Benkovic, S.J.; Huang, T.J. Lab-on-a-chip technologies for single-molecule studies. Lab. Chip 2013, 13, 2183–2198.
- Granéli, A.; Yeykal, C.C.; Prasad, T.K.; Greene, E.C. Organized arrays of individual DNA molecules tethered to supported lipid bilayers. Langmuir 2006, 22, 292–299.
- Fazio, T.; Visnapuu, M.-L.; Wind, S.; Greene, E.C. DNA curtains and nanoscale curtain rods: High-throughput tools for single molecule imaging. Langmuir 2008, 24, 10524–10531.
- Al Alwan, B.; AbuZineh, K.; Nozue, S.; Rakhmatulina, A.; Aldehaiman, M.; Al-Amoodi, A.S.; Serag, M.F.; Aleisa, F.A.; Merzaban, J.S.; Habuchi, S. Single-molecule imaging and microfluidic platform reveal molecular mechanisms of leukemic cell rolling. Commun. Biol. 2021, 4, 868.
- Zhang, C.; Jiang, K.; Liu, F.; Doyle, P.S.; van Kan, J.A.; van der Maarel, J.R. A nanofluidic device for single molecule studies with in situ control of environmental solution conditions. Lab. A Chip 2013, 13, 2821–2826.
More
