Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Catherine Yang and Version 1 by Atul Pandey.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused a global pandemic threat with more than 11.8 million confirmed cases and more than 0.5 million deaths as of 3 July 2020. Lung alveolar epithelial cells are considered as primary entry targets cells for the SARS-COV-2 through the angiotensin-converting enzyme-2 (ACE2) receptor, though it is expressed by numerous tissues. Both SARS-COV-2 and SARS-CoV utilize the ACE2 receptor to begin infection despite amino acid variation at a specific residue in the ACE2 receptor, suggesting that these variations might have been selected or could have increased the virulence and transmissibility of SARS-COV-2 compared to SARS-CoV.
- COVID-19
- adoptive cell-based therapy
- immunomodulatory drugs
1. Innate and Adaptive Immune Cells in COVID-19 Patients
SARS-CoV-2 infection activates both innate and adaptive immune response, where severe inflammatory response may cause local tissue damage (acute lung injury) and ARDS at systemic level [11,12][1][2]. Therefore, the knowledge behind this enhanced activation of cytokine storm due to dysregulated immune function after SARS-COV-2 infection will provide ways to clinically manage and prevent its transmission from mild to severe stage. Notably, bronchial mucosal-associated invariant T (MAIT) cells and γδ T cells are the primary innate immune cells that can trigger cytokines response after SARS-COV-2 infection, especially in patients developing the severe disease [13][3]. As a result of the activation of these innate immune cells and the consequential expression of pro-inflammatory cytokines genes, the host adaptive immune system becomes activated against virus infection.
Circulating white blood cells, including neutrophil, basophils, eosinophils numbers are consistently higher in survivors of COVID-19 than in non-survivors [14,15][4][5]. SARS-CoV-2 infection also induces lymphocytopenia in clinically severe patients that mostly affects the CD4+, CD8+ T cell subset, including effector, memory, regulatory T cells, and natural killer cells [11,12][1][2]. These observations are in line with the previous findings in severe or lethal cases of SARS-CoV and MERS [16,17][6][7]. This reduction in immune cells repertoire could be due to damage of lymphocytes or lymphocytic organs associated with infection with SARS-CoV-2 via their minimally expressed ACE2 receptors. Notably, the baseline levels of CD8+ T cells and NK cells are inversely correlated to ACE2 expression in human lung tissue [18][8]. Cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells are considered as a critical cell that controls viral infection. These cells were found to be exhausted in SARS-CoV-2-infected patients with a significant increase in the exhaustion marker PD1 (programmed death-1) compared to healthy controls and thus likely responsible for viral progression [19,20,21][9][10][11]. A recent study from Yang et al. demonstrated clinical outcomes of 93 SARS-CoV-2-positive patients in association with neutrophil(NEU)-to-lymphocyte (LYM) ratio (NLR), lymphocyte-to-monocyte (MON) ratio, platelet-to-lymphocyte ratio (PLR), and C-reactive protein (CRP) expression. According to their study, the aged patient group with elevated NLR was significantly associated with illness severity and could represent an independent prognostic biomarker for COVID-19 patients [22][12]. Similarly, another group in China with a cohort of 245 SARS-CoV-2-positive patients identified a patient group with elevated NLR, which was at a hyper-risk side compared to the other groups [23][13].
2. High-Throughput Sequencing Approach to Understand Immune Cell Dysfunctions in SARS-CoV-2-Infected Patients
Recently, Wen et al. have comprehensively characterized changes in transcriptional landscape during the recovery stage of SARS-CoV-2 infection by single-cell RNA sequencing (scRNA-seq) using peripheral blood mononuclear cells (PBMC) [24][14]. According to their study, inflammatory cytokines gene expressing CD14+ monocytes and plasma B-cell numbers were remarkably increased in COVID-19 patients during the early recovery stage [24][14]. Mingfeng et al. characterized bronchoalveolar lavage fluid (BALF)-immune cells through scRNA-seq from patients with varying severity of SARS-CoV-2 (mild and severe). BALF from critical/acute SARS-CoV-2-infected patients showed a higher proportion of macrophages and neutrophils and a lower proportion of myeloid/plasmacytoid dendritic cells and T cells compared with moderately infected patients [25][15]. Another study using scRNA-seq data identified specific cell types expressing a receptor for coronavirus infection (ACE2) across 13 tissue types; these include lung alveolar cells type-2, liver cholangiocyte, colon colonocytes, esophagus keratinocytes, stomach epithelial cells, and kidney proximal tubules [26][16]. Importantly, it has been observed that disease conditions, such as chronic heart disease or chronic cigarette smoke exposure, showed enhanced ACE2 expression notified by scRNA-seq data in cardiomyocytes or human lung cells, respectively [27,28][17][18]. Notably, human testicular cells (spermatogonia, Leydig, and Sertoli cells) also predominantly express the ACE2 receptor [29][19] representing that these tissue-specific cells are vulnerable to SARS-COV-2 infection [30][20]. It is important to note that not only over- or under-expression of ACE2 receptor in human tissue could determine the susceptibility of patient cells to SARS-COV-2, but also ACE2 polymorphism could influence both the predisposition to infection and clinical outcome of the COVID-19 pathogenesis [31][21]. Using mass-spectrometry, one study has profiled cellular immune components from SARS-CoV-2-infected patients with differences in disease progression (mild, severe, and critical) and compared with peripheral blood cells collected from healthy donors. According to their study, CD8+ T cells, dendritic cells, and macrophages were excessively activated initially during mild disease. They became exhausted in later critical stages, thereby representing disturbed homeostasis of the immune system during disease symptoms progression [32][22].
The scRNA-seq approach has also been utilized to identify novel therapeutic regimens for SARS-CoV-2 infection. In this line, a bioinformatics pipeline has been developed, which integrates the scRNA-seq dataset with a drug perturbation database to identify potential therapeutic candidates for SARS-CoV-2 infection treatment. Using bioinformatics pipeline, four drugs—(1) didanosine, an HIV anti-viral drug; (2) benzyl-quinazoline-4-yl-amine, an EGFR inhibitor; (3) camptothecin, a topoisomerase inhibitor; and (4) RO-90-7501, an amyloid-β42 aggregation inhibitor—have been proposed as potential candidates to treat COVID-19 [33][23]. Using knowledge from scRNA-seq and B-cell VDJ sequencing data, Cao et al. developed a therapeutic SARS-COV-2 neutralizing antibody (BD-368-2), with strong therapeutic and prophylactic efficacy in SARS-COV-2, and infected hACE2-transgenic mice [34][24]. Further, using cryoelectron microscopy, the structure of BD-368-2 in complex with spike-ectodomain trimer was characterized, which revealed BD-368-2 binding to the ACE2 receptor [34][24].
3. Hyper-Cytokine Activation
SARS-CoV-2 infection drives a profound cytokine response in the host, comprising a series of mediators that are targeted in immune-mediated inflammatory diseases (IMIDs) (Figure 1). In some patients, a condition of hypercytokinemia also called a cytokine storm with SARS-COV-2 infection develops, which resembles secondary hemophagocytic lymphohistiocytosis, a hyper-inflammatory state triggered by viral infections [35][25]. Recent reports have shown the level of plasma concentration of pro-inflammatory cytokines, such as TNF-α, IL-1β, IL-6, IL-8, IL-9, IL-10, bFGF, G-CSF, and GM-CSF, as well as chemokines, such as MCP1, IP10, and MIP1α, are elevated in patients with SARS-CoV-2 infection who are either admitted to an intensive care unit (ICU) or non-ICU patients compared to blood from healthy donors. These elevated cytokines levels were associated with lung injury [11,36][1][26]. Inflammatory cytokines were significantly elevated in patients who displayed severe clinical conditions and dismal clinical outcomes compared to moderately or mildly infected patients, thus suggesting that extensive changes in cytokines play a pivotal role in COVID-19 pathogenesis [15,37][5][27].
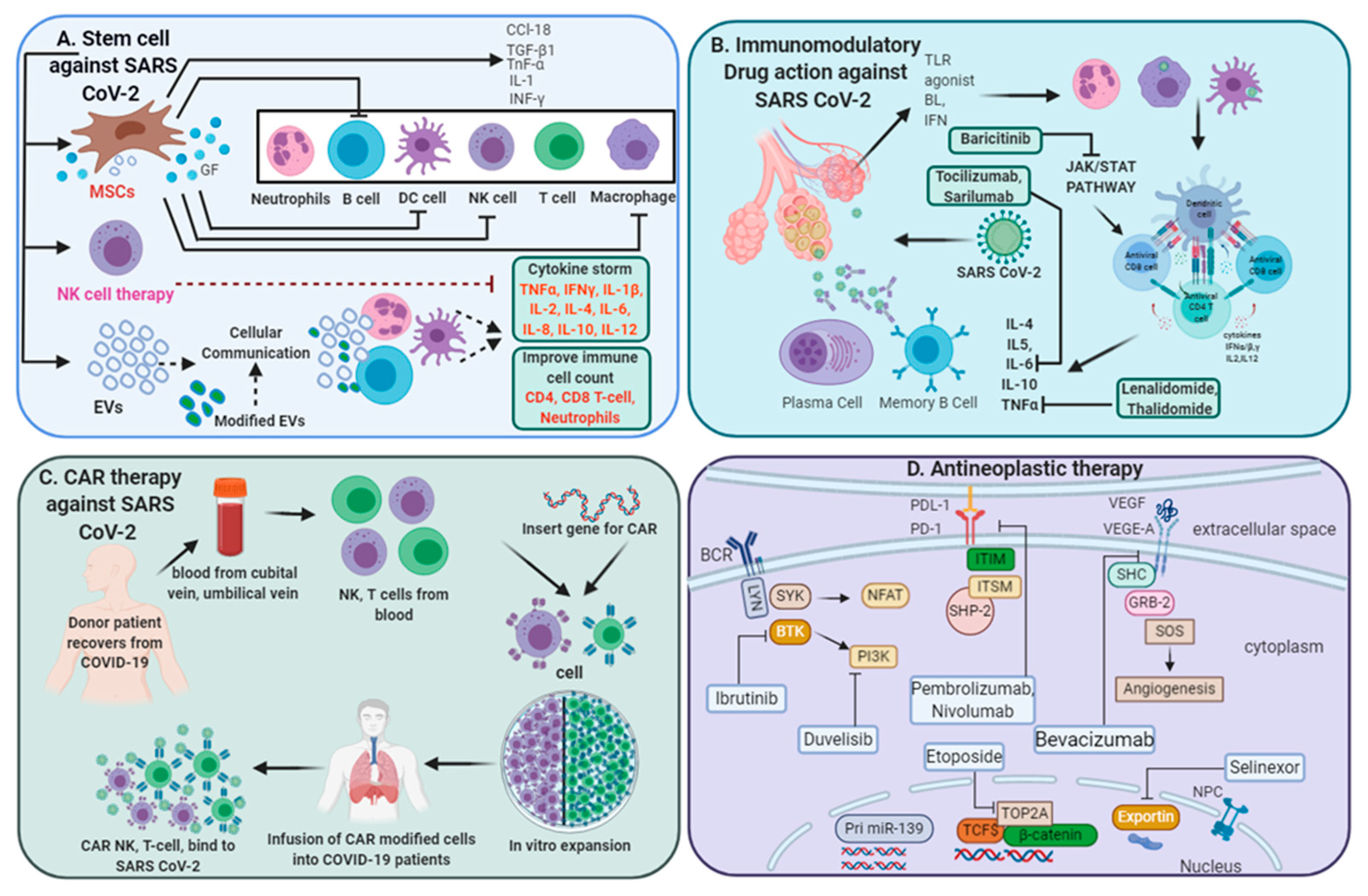
Figure 1. Multi-directive therapeutic intervention currently in clinical trials for combating SARS-CoV-2 pathogenesis. (A) Mesenchymal stem cell (MSC) and NK cell-based therapies for the treatment of SARS CoV-2 pathogenesis. Extracellular vesicles (EVs) derived from MSCs or modified exosomes containing antiviral/anti-inflammatory drugs or respective nucleic acid for therapeutic intervention to target cells. These cell-based or cell-derived therapeutic agents could modulate immune cell functions against SARS-CoV-2 infection. (B) Cytokine storm development is a central pandemic delinquent in SARS-CoV-2 infection. Certain immunomodulatory agents with excellent safety profiles or current anti-neoplastic interventions (D) may be considered for use in combination with antiviral drugs for the treatment of severe or critical COVID-19 cases. (C) Importantly, engineered immune cell receptors, including the chimeric antigen receptor (CAR) T-cell, NK cell therapy, offer new therapeutic approaches for SARS-CoV-2 infection. These immune cells collected from recovered patients offer an advantage as the majority of these cells have been primed with a viral antigen before collection and therefore transmit high proliferative efficiency and anti-viral efficacy.
4. SARS-COV-2 Severity in Cancer Patients
Besides the development of cytokine storm and exhaustion of immune cells, the clinical outcomes of SARS-CoV-2 infection are also dependent on multiple factors, such as age; clinical morbidities, including metabolic disorders like obesity, diabetes, cardiovascular and liver disease, and other conditions, such as pregnancy and cancer [38,39,40][28][29][30]. Notably, the expression of ACE2 receptors tend to increase with increasing age, and the majority of cancer is diagnosed at near to 60 years of age, marking cancer patients as more prone to SARS-CoV-2 infection. Consequently, that led to adverse clinical outcomes [41][31]. In addition to changes in the ACE2 receptor, immune functions in cancer patients are compromised. In fact, there is an increase in the expression of immunosuppressive cytokines and markers, such as PD1/PD-L1 on immune cells, which dampened the immune system and augmented the probability of viral infection [42][32]. Moreover, specific cytokines, such as IL17, secreted by Th17 cells in response to viral infection, have been shown to play a central role in virus pathogenesis through regulating the program cell death, cytokine storm, and lung cancer progression through VEGF (vascular endothelial growth factor) expression stimulation [40,43,44][30][33][34]. Therefore, therapies targeting such cytokines and anti-neoplastic agents might ideally improve the clinical efficacy of SARS-CoV-2-infected cancer patients [45][35].
5. Immune Responses of Asymptomatic Patients with SARS-CoV-2 Infection
Immune responses and clinical features of asymptomatic individuals with COVID-19 have not been well defined. These individuals comprise approximately 40–45% of the infected population and can silently spread the virus to others. The absence of symptoms in these infected patients does not mean that they are away from ultimate mortality risk, as they might have viral load equivalent to symptomatic patients. Therefore, more investigations are needed to understand the significant changes in disease symptoms, viral load, proper examination, and immune responses; all this knowledge might be useful to develop directed therapy to prevent the spread of this infection. Compared to asymptomatic people, recovered individuals (virus-negative/antibody-positive) can safely interact with susceptible and infected individuals [46][36]. Long Q and colleagues studied clinical features and immune responses of 37 asymptomatic individuals (20.8%), identified in a group of total 178 Real time polymerase chain reaction confirmed SARS-COV-2-positive people in the Wanzhou District of China [47][37]. However, this might not be an accurate assessment of the percentage of asymptomatic infections in the general population, since asymptomatic infections were identified from those who are at high risk of infection and not from a random sample of people. Therefore, the proportion of asymptomatic infections needs to be determined through population screening [48][38].
According to Long Q et al., the median duration of virus-shedding time in asymptomatic individuals was significantly longer than the symptomatic group. Moreover, the levels of virus-specific immunoglobulin (IgG) and levels of 18 anti-inflammatory cytokines were significantly lower in the asymptomatic group [47][37]. Previous studies have shown that circulating antibodies against SARS-CoV or MERS-CoV could remain for nearly 3 years or longer [49,50][39][40]. Several studies have reported that most SARS-CoV-2 convalescent individuals have detectable neutralizing antibodies, which correlate with the numbers of virus-specific T cells [51,52][41][42]. Additional longitudinal serological studies profiling more symptomatic and asymptomatic individuals are urgently needed to determine the duration of antibody-mediated immunity.
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506.
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422.
- Coperchini, F.; Chiovato, L.; Croce, L.; Magri, F.; Rotondi, M. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020, 53, 25–32.
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069.
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020.
- Perlman, S.; Dandekar, A.A. Immunopathogenesis of coronavirus infections: Implications for SARS. Nat. Rev. Immunol. 2005, 5, 917–927.
- Zumla, A.; Hui, D.S.; Perlman, S. Middle East respiratory syndrome. Lancet 2015, 386, 995–1007.
- Duijf, P.H.G. Baseline pulmonary levels of CD8+ T cells and NK cells inversely correlate with expression of the SARS-CoV-2 entry receptor ACE2. bioRxiv 2020.
- Zheng, M.; Gao, Y.; Wang, G.; Song, G.; Liu, S.; Sun, D.; Xu, Y.; Tian, Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol. Immunol. 2020, 17, 533–535.
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front. Immunol. 2020, 11, 827.
- Zhang, C.; Wang, X.M.; Li, S.R.; Twelkmeyer, T.; Wang, W.H.; Zhang, S.Y.; Wang, S.F.; Chen, J.Z.; Jin, X.; Wu, Y.Z.; et al. NKG2A is a NK cell exhaustion checkpoint for HCV persistence. Nat. Commun. 2019, 10, 1507.
- Yang, A.P.; Liu, J.P.; Tao, W.Q.; Li, H.M. The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int. Immunopharmacol. 2020, 84, 106504.
- Liu, Y.; Du, X.; Chen, J.; Jin, Y.; Peng, L.; Wang, H.H.X.; Luo, M.; Chen, L.; Zhao, Y. Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. J. Infect. 2020, 81, e6–e12.
- Wen, W.; Su, W.; Tang, H.; Le, W.; Zhang, X.; Zheng, Y.; Liu, X.; Xie, L.; Li, J.; Ye, J.; et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020, 6, 31.
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020, 26, 842–844.
- Qi, F.; Qian, S.; Zhang, S.; Zhang, Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020, 526, 135–140.
- Xu, D.; Ma, M.; Xu, Y.; Su, Y.; Ong, S.B.; Hu, X.; Chai, M.; Zhao, M.; Li, H.; Chen, Y.; et al. Single-cell Transcriptome Analysis Indicates New Potential Regulation Mechanism of ACE2 and NPs signaling among heart failure patients infected with SARS-CoV-2. medRxiv 2020.
- Smith, J.C.; Sausville, E.L.; Girish, V.; Yuan, M.L.; Vasudevan, A.; John, K.M.; Sheltzer, J.M. Cigarette Smoke Exposure and Inflammatory Signaling Increase the Expression of the SARS-CoV-2 Receptor ACE2 in the Respiratory Tract. Dev. Cell 2020, 53, 514–529.e513.
- Wang, Z.; Xu, X. scRNA-seq Profiling of Human Testes Reveals the Presence of the ACE2 Receptor, A Target for SARS-CoV-2 Infection in Spermatogonia, Leydig and Sertoli Cells. Cells 2020, 9, 920.
- Zou, X.; Chen, K.; Zou, J.; Han, P.; Hao, J.; Han, Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020, 14, 185–192.
- Devaux, C.A.; Rolain, J.M.; Raoult, D. ACE2 receptor polymorphism: Susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J. Microbiol. Immunol. Infect. 2020, 53, 425–435.
- Wang, W.; Su, B.; Pang, L.; Qiao, L.; Feng, Y.; Ouyang, Y.; Guo, X.; Shi, H.; Wei, F.; Su, X.; et al. High-dimensional immune profiling by mass cytometry revealed immunosuppression and dysfunction of immunity in COVID-19 patients. Cell Mol. Immunol. 2020, 17, 650–652.
- Alakwaa, F.M. Repurposing Didanosine as a Potential Treatment for COVID-19 Using Single-Cell RNA Sequencing Data. mSystems 2020, 5.
- Cao, Y.; Su, B.; Guo, X.; Sun, W.; Deng, Y.; Bao, L.; Zhu, Q.; Zhang, X.; Zheng, Y.; Geng, C.; et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell 2020.
- Pedersen, S.F.; Ho, Y.C. SARS-CoV-2: A storm is raging. J. Clin. Investig. 2020, 130, 2202–2205.
- Kabashneh, S.; Ali, H.; Alkassis, S. Multi-Organ Failure in a Patient With Diabetes due to COVID-19 With Clear Lungs. Cureus 2020, 12, e8147.
- Alijotas-Reig, J.; Esteve-Valverde, E.; Belizna, C.; Selva-O’Callaghan, A.; Pardos-Gea, J.; Quintana, A.; Mekinian, A.; Anunciacion-Llunell, A.; Miro-Mur, F. Immunomodulatory therapy for the management of severe COVID-19. Beyond the anti-viral therapy: A comprehensive review. Autoimmun. Rev. 2020, 19, 102569.
- Costa, F.F.; Rosario, W.R.; Ribeiro Farias, A.C.; de Souza, R.G.; Duarte Gondim, R.S.; Barroso, W.A. Metabolic syndrome and COVID-19: An update on the associated comorbidities and proposed therapies. Diabetes Metab. Syndr. 2020, 14, 809–814.
- Dashraath, P.; Wong, J.L.J.; Lim, M.X.K.; Lim, L.M.; Li, S.; Biswas, A.; Choolani, M.; Mattar, C.; Su, L.L. Coronavirus disease 2019 (COVID-19) pandemic and pregnancy. Am. J. Obs. Gynecol. 2020, 222, 521–531.
- Allegra, A.; Pioggia, G.; Tonacci, A.; Musolino, C.; Gangemi, S. Cancer and SARS-CoV-2 Infection: Diagnostic and Therapeutic Challenges. Cancers 2020, 12, 1581.
- Gosain, R.; Abdou, Y.; Singh, A.; Rana, N.; Puzanov, I.; Ernstoff, M.S. COVID-19 and Cancer: A Comprehensive Review. Curr. Oncol. Rep. 2020, 22, 53.
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science 2011, 331, 1565–1570.
- Pan, B.; Che, D.; Cao, J.; Shen, J.; Jin, S.; Zhou, Y.; Liu, F.; Gu, K.; Man, Y.; Shang, L.; et al. Interleukin-17 levels correlate with poor prognosis and vascular endothelial growth factor concentration in the serum of patients with non-small cell lung cancer. Biomarkers 2015, 20, 232–239.
- Patera, A.C.; Pesnicak, L.; Bertin, J.; Cohen, J.I. Interleukin 17 modulates the immune response to vaccinia virus infection. Virology 2002, 299, 56–63.
- Zumla, A.; Hui, D.S.; Azhar, E.I.; Memish, Z.A.; Maeurer, M. Reducing mortality from 2019-nCoV: Host-directed therapies should be an option. Lancet 2020, 395, e35–e36.
- Weitz, J.S.; Beckett, S.J.; Coenen, A.R.; Demory, D.; Dominguez-Mirazo, M.; Dushoff, J.; Leung, C.Y.; Li, G.; Magalie, A.; Park, S.W.; et al. Modeling shield immunity to reduce COVID-19 epidemic spread. Nat. Med. 2020, 26, 849–854.
- Long, Q.X.; Tang, X.J.; Shi, Q.L.; Li, Q.; Deng, H.J.; Yuan, J.; Hu, J.L.; Xu, W.; Zhang, Y.; Lv, F.J.; et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020, 26, 1200–1204.
- Gudbjartsson, D.F.; Helgason, A.; Jonsson, H.; Magnusson, O.T.; Melsted, P.; Norddahl, G.L.; Saemundsdottir, J.; Sigurdsson, A.; Sulem, P.; Agustsdottir, A.B.; et al. Spread of SARS-CoV-2 in the Icelandic Population. N. Engl. J. Med. 2020, 382, 2302–2315.
- Cao, W.C.; Liu, W.; Zhang, P.H.; Zhang, F.; Richardus, J.H. Disappearance of antibodies to SARS-associated coronavirus after recovery. N. Engl. J. Med. 2007, 357, 1162–1163.
- Choe, P.G.; Perera, R.; Park, W.B.; Song, K.H.; Bang, J.H.; Kim, E.S.; Kim, H.B.; Ko, L.W.R.; Park, S.W.; Kim, N.J.; et al. MERS-CoV Antibody Responses 1 Year after Symptom Onset, South Korea, 2015. Emerg. Infect. Dis. 2017, 23, 1079–1084.
- Thevarajan, I.; Nguyen, T.H.O.; Koutsakos, M.; Druce, J.; Caly, L.; van de Sandt, C.E.; Jia, X.; Nicholson, S.; Catton, M.; Cowie, B.; et al. Breadth of concomitant immune responses prior to patient recovery: A case report of non-severe COVID-19. Nat. Med. 2020, 26, 453–455.
- Suthar, M.S.; Zimmerman, M.; Kauffman, R.; Mantus, G.; Linderman, S.; Vanderheiden, A.; Nyhoff, L.; Davis, C.; Adekunle, S.; Affer, M.; et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. medRxiv 2020.
More
