Cotton fiber development largely depends on cell wall biosynthesis and cytoskeleton arrangement. Cytoskeleton dynamics control many cellular processes, such as the movement of organelles, cell wall formation, and cell division. Microfilaments (actin-filament), microtubules, and intermediate filaments are the main constituents of the cytoskeleton. In most cells, actin filaments are involved in secretory vesicle transportation to the cell membrane and cell wall, enhancing cell expansion. The actin cytoskeleton also regulates tip growth and cell elongation. Dozens express actin proteins to hundreds of genes in the ACTIN family. Arabidopsis has 10 actin genes, of which 8 are functional, and 2 are categorized as pseudogenes, while cotton plants have been identified with 16 actin genes.
1. Actin Filament Development Pathway and Actin-Binding Proteins (ABPs)
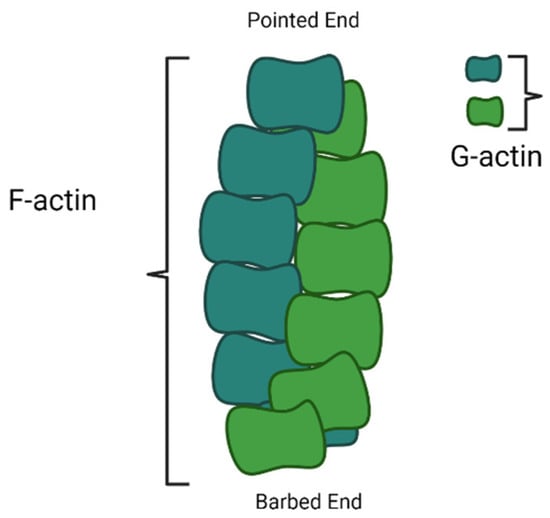
Actin is expressed in monomeric form, which is known as G-actin. The G-actin polymerises form a filament known as F-actin
[1][58]. The formation of actin filaments by monomeric actin includes nucleation, polymerization/capping, and F-actin bundling and cross-linking activities. Many Actin-Binding Proteins (ABPs) are divided according to their association among G-Actin-binding/G-actin capping proteins and F-Actin regulators (ABPs), which are involved in either polymerization or depolymerization, and proteins that serve to crosslink and/or bundle the actin microfilaments
[2][59]. The detailed pathway and the ABPs are involved in every step of actin microfilament formation.
2. Nucleation of Actin Filaments
Nucleation is the first step in the formation of a new filament. The nucleation of actin microfilaments is not a spontaneous process; rather, it depends on many nucleating proteins. Polymerization is energetically hostile unless actin monomers are nucleated. During polymerization/capping, actin filaments form two ends; a pointed end, which is slow growing, and a barbed end, which is fast growing. Arp2/3 complex is one of the first known nucleating molecules, which caps at a pointed end and makes the barbed end available for polymerization. However, WASP homology2 domains of WASP protein, SCAR-WAVE protein, and verprolin/WIP have also been very important for the Arp2/3 complex in actin nucleation
[2][3][59,60]. Foramins have also been found in actin filament nucleation, apart from the Arp2/3 complex. The formin proteins and Spire also nucleate actin polymerization. However, studies indicated that the foramin homology2 (FH2) domain dimer stretches to accommodate the progressive addition of actin monomers to the barbed end of a filament
[4][61].
3. Polymerization and Capping of Microfilament
After nucleation, the microfilaments start growing by adding actin monomers at the barbed end, also known as the growing end; however, the pointed end is a non-growing end (
Figure 1). The FH2 domain of formins directly nucleates actin monomers to form actin filaments by protecting growing ends from capping proteins while guiding the rapid insertion of new actin subunits. Residues Ile1431 in the knob and Lys1601 and Lys1359 in the post of the FH2 domain of Bni1p, a formin protein, manifest as actin-binding sites. The FH1 domain of formins not only recruits profilin–actin complexes but also accelerates filament elongation at least five times faster than the rate of diffusion-limited subunit addition at the free barbed ends of filaments
[5][62]. The process of actin filament development happens at the non-growing pointed end.
Figure 1. Diagram showing polymerization of F-actin filament on barded end (image drawn by the authors).
Once the filaments grow enough, the length is controlled by many proteins, usually called cappers proteins, such as gelsolins and tensins, which inhibit the addition of further monomers. Adding cappers proteins at the pointed ends reduces the monomer loss and facilitates the extension of microfilament
[6][63].
The polymerized filament (F-actin) can be depolymerized by Actin Depolymerized Factor (ADF)/Cofilin. The depolymerization activity of ADF/Cofilin complex is further enhanced by Actin Interaction Protein-1 (AIP-1)
[2][59]. In contrast to the filament, the depolymerization protein, tropomyosin, and nubuline have been identified to stabilize the actin filament in muscle cells. Several other proteins have poly-proline-motifs filament stabilization by recruiting polymerization machinery
[4][61].
4. F-Actin Bundling and Cross Linking
After actin F-filament formation, the next step is actin bundling which is carried out by the alignment of the F-filament (F-actin) in a parallel or anti-parallel manner. Actin filament bundling is usually accomplished by proteins with two actin-binding domains
[4][61]. The arrangement of bundled actin-filaments into orthogonal arrays is further mediated by proteins having multi-actin-binding domains. Actin-binding proteins, involved in cross-linking processes, have two or many domains, usually separated by a spacer. Filamin (dimeric) or spectrin (tetrameric) proteins cross-link. A monomeric protein called tansgelin has also been reported to be involved in cross-linking
[7][64].
5. Plant LIM, an Actin-Bundling Protein
Plant LIM proteins are another important class of ABPs
[8][65], which are found to be dispersed in the cytosol and nucleoplasm. The LIM-domain-containing proteins in the nucleus are preferentially involved in tissue-specific gene regulation and determination of cell fate, whereas the cytoplasmic LIM-domain-containing proteins are involved mainly in cytoskeletal organization
[9][66]. The term “LIM” originates from the initials LIN-11, ISL-1, and MEC-3, the first proteins observed to contain this particular homeodomain. Following this, all proteins containing LIM domains are called LIM proteins or LIM domain-containing proteins
[10][67]. Most of the LIM proteins have two different LIM domains, each comprising 55 amino acids
[11][68] and having the broad consensus sequence (CX2CX16-23HX2C)X2(CX2CX15-30CX2C/H/D) in which eight cys-his conserved residues form two zinc finger projections. The motifs in LIM domains are involved in protein–protein interactions and possess conserved scaffolds that recognize a diverse variety of target proteins. Each Zinc-finger motif within the LIM domains contains two Zinc coordinating cys-residues which assist in forming a β hairpin connection with the target protein. In LIM2, the single LIM domain consists of two Zinc fingers with a core of bulky hydrophobic residues
[12][69]. Phylogenetic analyses of plant LIM proteins separate them into seven classes (XLIM1, WLIM1, WLIM2, βLIM1, PLIM1, PLIM2, and PLIM2-like)
[13][70]; or into six categories, in which GhLIM1, GhWLIM2, and GhWLIM5 belong to the WLIM2 subgroup. Bioinformatics analysis shows that GhWLIM2 and GhWLIM5 have strong evolutionary relationships
[14][41]. In cotton, there are many LIM-domain-containing proteins that modulate actin filament bundlings, such as GhPLIM1, which is predominantly involved in anther development
[10][67], and WLIM1a, which is involved in fiber elongation along with secondary wall synthesis
[15][49]. Cotton WLIM1a contains two domains: Domain 1 (D1) is involved in actin-bundling activity, whereas Domain 2 (D2) participates in DNA binding
[8][65].
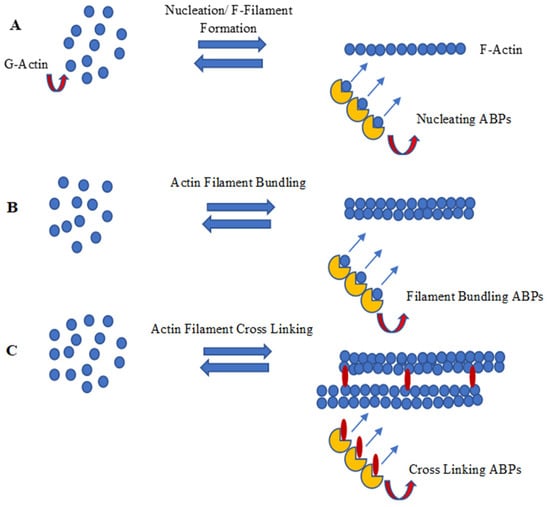
Figure 2 summarizes nucleation, polymerization/capping, and F-actin bundling and the cross-linking process of actin filament development through a schematic diagram.
Figure 2. Schematic diagram of (A) nucleation, (B) polymerization/capping and (C) F-actin bundling and cross linking (image drawn by the researcheuthors).