Cancer is one of the leading causes of death worldwide, and its incidence and mortality are increasing each year. Improved therapeutic strategies against cancer have progressed, but remain insufficient to invert this trend. Along with several other risk factors, abnormal genetic and epigenetic regulations play a critical role in the initiation of cellular transformation, as well as tumorigenesis. The epigenetic regulator UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1) is a multidomain protein with oncogenic abilities overexpressed in most cancers. Through the coordination of its multiple domains and other epigenetic key players, UHRF1 regulates DNA methylation and histone modifications. This well-coordinated dialogue leads to the silencing of tumor-suppressor genes (TSGs) and facilitates tumor cells’ resistance toward anticancer drugs, ultimately promoting apoptosis escape and uncontrolled proliferation. Several studies have shown that the downregulation of UHRF1 with natural compounds in tumor cells induces the reactivation of various TSGs, inhibits cell growth, and promotes apoptosis.Cancer is one of the leading causes of death worldwide, and its incidence and mortality are increasing each year. Improved therapeutic strategies against cancer have progressed, but remain insufficient to invert this trend. Along with several other risk factors, abnormal genetic and epigenetic regulations play a critical role in the initiation of cellular transformation, as well as tumorigenesis. The epigenetic regulator UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1) is a multidomain protein with oncogenic abilities overexpressed in most cancers. Through the coordination of its multiple domains and other epigenetic key players, UHRF1 regulates DNA methylation and histone modifications. This well-coordinated dialogue leads to the silencing of tumor-suppressor genes (TSGs) and facilitates tumor cells’ resistance toward anticancer drugs, ultimately promoting apoptosis escape and uncontrolled proliferation. Several studies have shown that the downregulation of UHRF1 with natural compounds in tumor cells induces the reactivation of various TSGs, inhibits cell growth, and promotes apoptosis. This entry discusses the underlying mechanisms and the potential of various natural and synthetic compounds that can inhibit/minimize UHRF1’s oncogenic activities and/or its expression.
- cancer
- DNA methylation
- epidrugs
- epigenetics
- phytochemical
1. Introduction
2. Structure and Function of UHRF1
2.1. Structure of UHRF1
Human UHRF1 (initially known as ICBP90) was discovered by two of us as a transcriptional regulator of topoisomerase IIα by binding to an inverted CCAAT box in its promoter region [7][8]. However, it is now well-characterized for its involvement in various epigenetic and cellular pathways, including the maintenance of methylation patterns on DNA, histone modifications, DNA damage repair, and the regulation of other proteins [9][10][11]. UHRF1 is a 793-amino-acid-long protein coded by the UHRF1 gene mapped at the 19p13.3 location in the human genome (Figure 1). UHRF1 is a protein with multiple domains that differ in their structure and functions [12][13]. Starting from the N-terminus, the domains are the ubiquitin-like domain (UBL), tandem Tudor domain (TTD), plant homeodomain (PHD), set and ring-associated (SRA) domain, and, finally, the really interesting new gene (RING) domain at the C-terminus of the protein (Figure 1).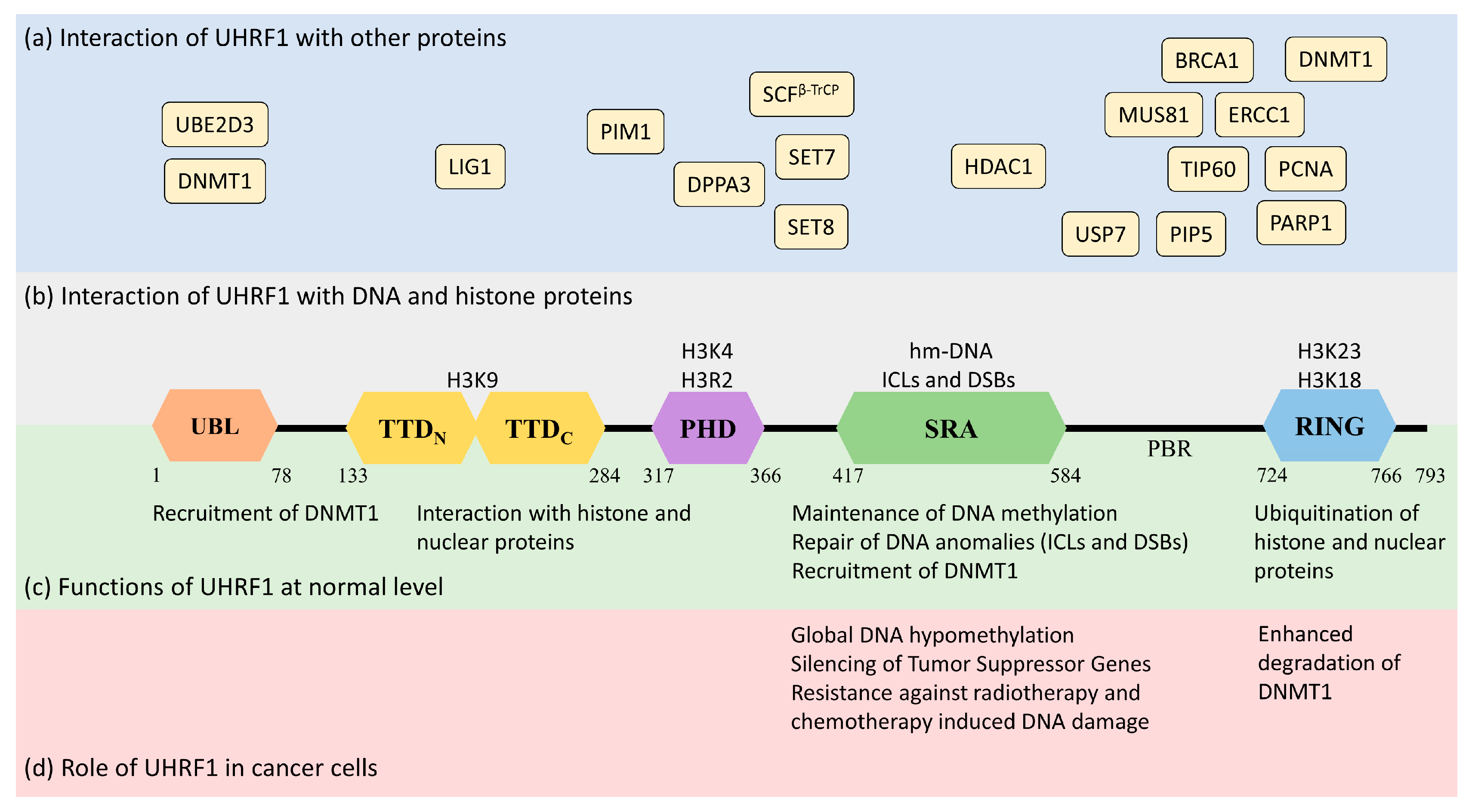
2.1.1. Ubiquitin-like Domain (UBL)
The UBL is structurally 35% identical to ubiquitin and is linked to the stability of UHRF1. It possesses characteristic α-helix and β-sheets, as well as two conserved lysine residues, K31 and K50 (similar to ubiquitin K29 and K48), which can be polyubiquitinated to trigger proteasomal degradation [9]. A role of the UBL has also been identified in DNMT1 (DNA methyltransferase 1) recruitment and the accurate transmission of methylation patterns [13]. Indeed, the UBL coordinates with the UHRF1 RING domain to ubiquitinate histone H3 residues, which serve as an anchorage signal for DNMT1 recruitment on hemi-methylated DNA [15][16]. Interestingly, the direct interaction of UHRF1-UBL with the RFTS (Replication Focus-Targeting Sequence) domain of DNMT1 also facilitates the enzymatic activity of the latter by activating its catalytic domain [17].2.1.2. Tandem Tudor Domain (TTD)
The Tandem Tudor domain of UHRF1 is involved in various protein–protein interactions vital for UHRF1 biological functions. The TTD notably allows UHRF1 to interact with the histone marks needed for its functioning [18]. The TTD is made up of two subdomains (TTDN and TTDC), each having a characteristic five-stranded β-barrel moiety in its structure [19]. The aromatic cage (Phe-152, Tyr-188, and Tyr-191) in TTDN, along with Asn-194 and Asp-145, recognizes di- and tri-methylated H3K9, while the peptide-binding groove between the TTDN and TTDC is sensitive to the epigenetic modifications on adjacent histone residues (i.e., H3K4 methylation and H3T6 phosphorylation) [19]. Through its interaction with LIG1 (DNA ligase), the TTD allows UHRF1 recruitment to replication foci to play its role in DNA methylation [20], but this interaction is not essential for abnormal DNA methylation patterns in cancerous cells [21]. The TTD also interacts with LIG1K126me3, which opens the closed conformation of UHRF1 [22][23]. In contrast, when the TTD interacts with the polybasic region (PBR) of UHRF1, the binding of H3K9me3 with the TTD-PHD domains is disrupted, ultimately returning to the closed conformation of UHRF1. Finally, when UHRF1 binds to hemi-methylated DNA or USP7 (Ubiquitin-Specific-processing Protease 7), the interaction of the TTD with the PBR is disturbed, which favors the transition to the open conformation of UHRF1 [24][25][26].2.1.3. Plant Homeodomain (PHD)
The plant homeodomain (PHD) of UHRF1 differs from the canonical PHDs in other proteins by having a PHD motif containing four cysteines (C302, C305, C313, and C316) in a loop coordinated with a zinc atom linked to the canonical PHD region by a single helical turn. The PHD domain of UHRF1 specifically recognizes the H3R2 motif in chromatin, which is essential for UHRF1 functions [27][28]. This interaction of the PHD with histone proteins can be altered through other proteins, such as DPPA3 (developmental pluripotency-associated protein 3), inhibiting UHRF1 localization on chromatin and promoting passive demethylation [29].2.1.4. Set and Ring-Associated (SRA) Domain
The SRA is a highly conserved domain specific to the UHRF family of proteins [9], playing an important role in DNA methylation [30][31][32]. Through this domain, UHRF1 recognizes hemi-methylated DNA and flips the methylated cytosine out of the helix. The SRA domain functions as a palm of a hand grasping the DNA duplex, where the NKR finger (489–491 amino acid residues) and thumb (444–496 amino acid residues) form two specific loops that project into major and minor grooves of the DNA double helix to read the nucleotides in the CpG duplex. The NKR finger specifically identifies the hemi-methylated DNA and flips the methylated cytosine out of the duplex. The flipped methylated cytosine is later stabilized by π-π stacking with the conserved tyrosine (466 and 478) residues in the binding domain [30]. The NKR domain also helps UHRF1 to differentiate between hemi-methylated and fully methylated DNA, since the second methylated cytosine creates a steric hindrance for the NKR finger, leading to a reduced affinity of UHRF1 for methylated DNA [30]. The only other protein from vertebrates carrying an SRA domain is UHRF2 [8]. In contrast to UHRF1, UHRF2 exhibits tumor-suppressor gene capacities [8][14], but an ability to favor tumor progression cannot be excluded, at least in certain types of cancer, such as hepatocellular carcinoma [33] or intestinal tumorigenesis [34]. The homology of amino-acid sequences between UHRF1 and UHRF2 reaches 74% [8][14], questioning the possibility that a drug targeting the SRA domain of UHRF1 putatively also may bind to the SRA domain of UHRF2 and thus may have opposite pharmacological effects. This seems, however, unlikely, but not impossible, considering the structures of each SRA domain. First, the UHRF2 SRA domain shows preferential recognition of hydroxymethyl cytosine over methylcytosine [35], suggesting differences in the respective structures. Indeed, the NKR loop, involved in the base-flipping mechanism, is disordered in the UHRF2-SRA domain, while in the UHRF1-SRA domain, it is not. This difference has been proposed to explain why the UHRF2-SRA domain has a preference for fully hydroxymethylated DNA over hemi-hydroxymethylated DNA vs. the UHRF1-SRA domain having a preference for hemi-methylated DNA over fully methylated DNA [35]. Altogether, this supports the fact that drugs targeting the SRA domains of UHRF1 would have specificity versus UHRF2, despite their strong similarities (77% amino acid sequence identity, personal observations).2.1.5. RING Domain
The RING domain harbors the only enzymatic activity of this protein [36]. It is rich in cysteine residues that form two zinc fingers interacting with a variety of substrates. Through this domain, UHRF1 can ubiquitinate itself, but also DNMT1, H3, and other proteins, regulating their function and stability [16][37].2.2. Functions of UHRF1
UHRF1 is an important component of an epigenetic complex acting after DNA replication. UHRF1 is primarily involved in the maintenance of genomic DNA methylation patterns in cells [9][38]. Via its SRA domain, UHRF1 recognizes the CpG motifs on the parent strand of hemi-methylated DNA and flips the methylated cytosine out of the duplex. This then enables DNMT1 to methylate the opposite cytosine on the newly formed daughter strand [30][31][32][39]. UHRF1 also helps in recruiting DNMT1 to the hemi-methylation sites by direct interaction through its UBL and SRA domains, or indirectly by ubiquitinating histone H3K18 through its RING domain, as H3K18ub serves as a binding site for DNMT1. The crosstalk of UHRF1 TTD with H3K9me3 and H3K4 or PHD with H3R2 also plays a significant role in the recognition of the methylation site and the maintenance of the methylation patterns on the daughter strand [19][40][41][42]. The recruitment and activity of UHRF1 at the replication site is also facilitated by the DNA replication machinery. DNA ligase 1 (LIG1), methylated by G9a and GLP methyltransferases, mimics H3K9me2/3 in binding to the TTD of UHRF1. This binding recruits UHRF1 to the DNA replication sites for the maintenance of DNA methylation [20]. Similarly, the interaction with hemi-methylated DNA, USP7, and PIP5 (Phosphatidylinositol-4-phosphate 5) during the S-phase, triggers UHRF1 to switch from its “closed” to its “open” state, which facilitates UHRF1 loading onto the newly formed DNA [24][26][43] (Figure 2).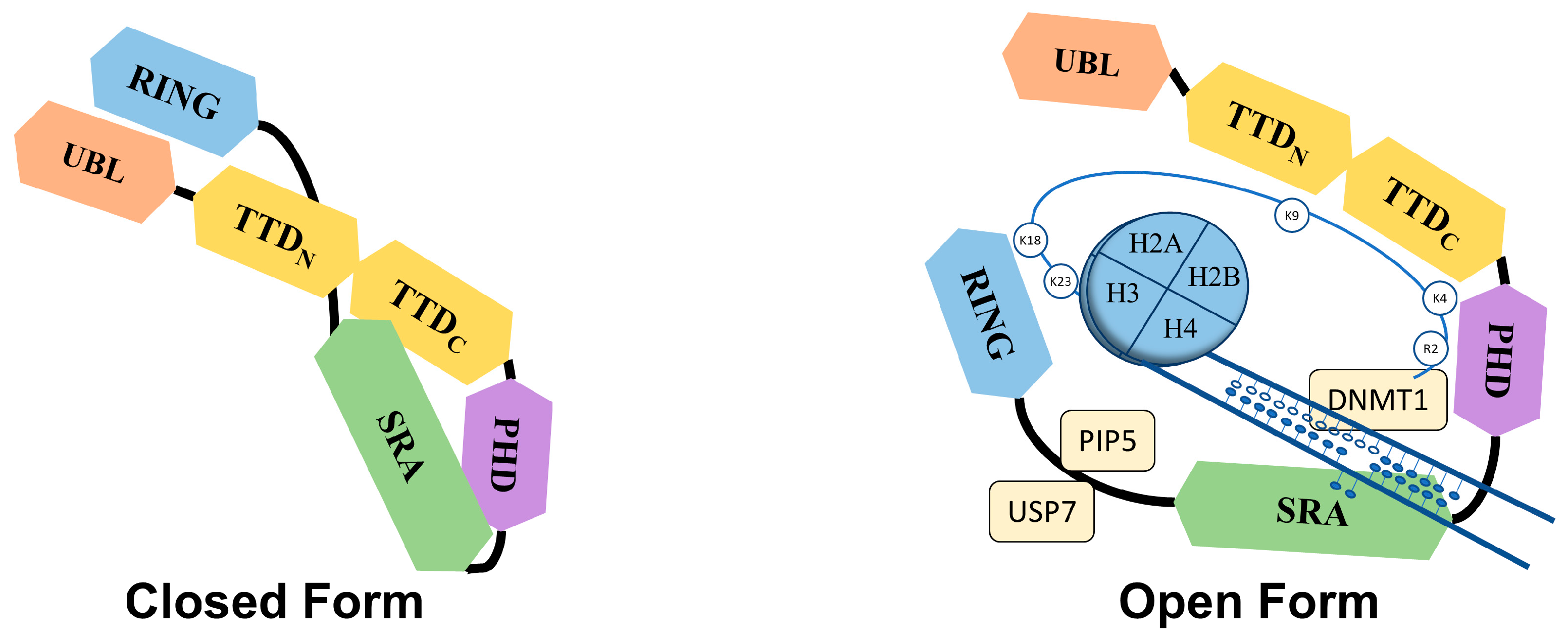
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424.
- You, W.; Henneberg, M. Cancer incidence increasing globally: The role of relaxed natural selection. Evol. Appl. 2017, 11, 140–152.
- Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 2008, 358, 1148–1159.
- Dupont, C.; Armant, D.R.; Brenner, C.A. Epigenetics: Definition, Mechanisms and Clinical Perspective; Thieme Medical Publishers: New York, NY, USA, 2009; Volume 27, pp. 351–357.
- Lund, A.H.; van Lohuizen, M. Epigenetics and Cancer. Genes Dev. 2004, 18, 2315–2335.
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36.
- Hopfner, R.; Mousli, M.; Garnier, J.-M.; Redon, R.; du Manoir, S.; Chatton, B.; Ghyselinck, N.; Oudet, P.; Bronner, C. Genomic structure and chromosomal mapping of the gene coding for ICBP90, a protein involved in the regulation of the topoisomerase IIα gene expression. Gene 2001, 266, 15–23.
- Hopfner, R.; Mousli, M.; Jeltsch, J.M.; Voulgaris, A.; Lutz, Y.; Marin, C.; Bellocq, J.P.; Oudet, P.; Bronner, C. ICBP90, a Novel Human CCAAT Binding Protein, Involved in the Regulation of Topoisomerase IIalpha Expression. Cancer Res. 2000, 60, 121–128.
- Bronner, C.; Achour, M.; Arima, Y.; Chataigneau, T.; Saya, H.; Schini-Kerth, V.B. The UHRF family: Oncogenes that are drugable targets for cancer therapy in the near future? Pharmacol. Ther. 2007, 115, 419–434.
- Mancini, M.; Magnani, E.; Macchi, F.; Bonapace, I.M. The multi-functionality of UHRF1: Epigenome maintenance and preservation of genome integrity. Nucleic Acids Res. 2021, 49, 6053–6068.
- Bronner, C.; Krifa, M.; Mousli, M. Increasing role of UHRF1 in the reading and inheritance of the epigenetic code as well as in tumorogenesis. Biochem. Pharmacol. 2013, 86, 1643–1649.
- Bronner, C.; Alhosin, M.; Hamiche, A.; Mousli, M. Coordinated Dialogue between UHRF1 and DNMT1 to Ensure Faithful Inheritance of Methylated DNA Patterns. Genes 2019, 10, 65.
- Smets, M.; Link, S.; Wolf, P.; Schneider, K.; Solis, V.; Ryan, J.; Meilinger, D.; Qin, W.; Leonhardt, H. DNMT1 mutations found in HSANIE patients affect interaction with UHRF1 and neuronal differentiation. Hum. Mol. Genet. 2017, 26, 1522–1534.
- Unoki, M.; Sasaki, H. The UHRF protein family in epigenetics, development, and carcinogenesis. Proc. Jpn. Acad. Ser. B 2022, 98, 401–415.
- DaRosa, P.A.; Harrison, J.S.; Zelter, A.; Davis, T.N.; Brzovic, P.; Kuhlman, B.; Klevit, R.E. A Bifunctional Role for the UHRF1 UBL Domain in the Control of Hemi-methylated DNA-Dependent Histone Ubiquitylation. Mol. Cell 2018, 72, 753–765.e6.
- Foster, B.M.; Stolz, P.; Mulholland, C.B.; Montoya, A.; Kramer, H.; Bultmann, S.; Bartke, T. Critical Role of the UBL Domain in Stimulating the E3 Ubiquitin Ligase Activity of UHRF1 toward Chromatin. Mol. Cell 2018, 72, 739–752.e9.
- Li, T.; Wang, L.; Du, Y.; Xie, S.; Yang, X.; Lian, F.; Zhou, Z.; Qian, C. Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation. Nucleic Acids Res. 2018, 46, 3218–3231.
- Lu, R.; Wang, G.G. Tudor: A versatile family of histone methylation ‘readers’. Trends Biochem. Sci. 2013, 38, 546–555.
- Nady, N.; Lemak, A.; Walker, J.R.; Avvakumov, G.V.; Kareta, M.S.; Achour, M.; Xue, S.; Duan, S.; Allali-Hassani, A.; Zuo, X.; et al. Recognition of Multivalent Histone States Associated with Heterochromatin by UHRF1 Protein. J. Biol. Chem. 2011, 286, 24300–24311.
- Ferry, L.; Fournier, A.; Tsusaka, T.; Adelmant, G.; Shimazu, T.; Matano, S.; Kirsh, O.; Amouroux, R.; Dohmae, N.; Suzuki, T.; et al. Methylation of DNA Ligase 1 by G9a/GLP Recruits UHRF1 to Replicating DNA and Regulates DNA Methylation. Mol. Cell 2017, 67, 550–565.e5.
- Vaughan, R.M.; Kupai, A.; Foley, C.A.; Sagum, C.A.; Tibben, B.M.; Eden, H.E.; Tiedemann, R.L.; Berryhill, C.A.; Patel, V.; Shaw, K.M.; et al. The histone and non-histone methyllysine reader activities of the UHRF1 tandem Tudor domain are dispensable for the propagation of aberrant DNA methylation patterning in cancer cells. Epigenetics Chromatin 2020, 13, 44.
- Jeltsch, A. Novel Insights into Peptide Binding and Conformational Dynamics of UHRF1. Structure 2019, 27, 408–410.
- Kori, S.; Ferry, L.; Matano, S.; Jimenji, T.; Kodera, N.; Tsusaka, T.; Matsumura, R.; Oda, T.; Sato, M.; Dohmae, N.; et al. Structure of the UHRF1 Tandem Tudor Domain Bound to a Methylated Non-histone Protein, LIG1, Reveals Rules for Binding and Regulation. Structure 2019, 27, 485–496.e7.
- Fang, J.; Cheng, J.; Wang, J.; Zhang, Q.; Liu, M.; Gong, R.; Wang, P.; Zhang, X.; Feng, Y.; Lan, W.; et al. Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition. Nat. Commun. 2016, 7, 11197.
- Gao, L.; Tan, X.-F.; Zhang, S.; Wu, T.; Zhang, Z.-M.; Ai, H.-W.; Song, J. An Intramolecular Interaction of UHRF1 Reveals Dual Control for Its Histone Association. Structure 2018, 26, 304–311.e3.
- Zhang, Z.-M.; Rothbart, S.B.; Allison, D.F.; Cai, Q.; Harrison, J.S.; Li, L.; Wang, Y.; Strahl, B.D.; Wang, G.G.; Song, J. An Allosteric Interaction Links USP7 to Deubiquitination and Chromatin Targeting of UHRF1. Cell Rep. 2015, 12, 1400–1406.
- Hu, L.; Li, Z.; Wang, P.; Lin, Y.; Xu, Y. Crystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2. Cell Res. 2011, 21, 1374–1378.
- Rajakumara, E.; Wang, Z.; Ma, H.; Hu, L.; Chen, H.; Lin, Y.; Guo, R.; Wu, F.; Li, H.; Lan, F.; et al. PHD Finger Recognition of Unmodified Histone H3R2 Links UHRF1 to Regulation of Euchromatic Gene Expression. Mol. Cell 2011, 43, 275–284.
- Hata, K.; Kobayashi, N.; Sugimura, K.; Qin, W.; Haxholli, D.; Chiba, Y.; Yoshimi, S.; Hayashi, G.; Onoda, H.; Ikegami, T.; et al. Structural basis for the unique multifaceted interaction of DPPA3 with the UHRF1 PHD finger. Nucleic Acids Res. 2022, 50, 12527–12542.
- Avvakumov, G.V.; Walker, J.R.; Xue, S.; Li, Y.; Duan, S.; Bronner, C.; Arrowsmith, C.H.; Dhe-Paganon, S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 2008, 455, 822–825.
- Arita, K.; Ariyoshi, M.; Tochio, H.; Nakamura, Y.; Shirakawa, M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 2008, 455, 818–821.
- Hashimoto, H.; Horton, J.R.; Zhang, X.; Bostick, M.; Jacobsen, S.E.; Cheng, X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 2008, 455, 826–829.
- Sun, J.; Wu, K.; Chen, S.; Jiang, S.; Chen, Y.; Duan, C. UHRF2 Promotes Hepatocellular Carcinoma Progression by Upregu-lating ErbB3/Ras/Raf Signaling Pathway. Int. J. Med. Sci. 2021, 18, 3097.
- Li, L.; Duan, Q.; Zeng, Z.; Zhao, J.; Lu, J.; Sun, J.; Zhang, J.; Siwko, S.; Wong, J.; Shi, T.; et al. UHRF2 promotes intestinal tumorigenesis through stabilization of TCF4 mediated Wnt/β-catenin signaling. Int. J. Cancer 2020, 147, 2239–2252.
- Zhou, T.; Xiong, J.; Wang, M.; Yang, N.; Wong, J.; Zhu, B.; Xu, R.-M. Structural Basis for Hydroxymethylcytosine Recognition by the SRA Domain of UHRF2. Mol. Cell 2014, 54, 879–886.
- Jenkins, Y.; Markovtsov, V.; Lang, W.; Sharma, P.; Pearsall, D.; Warner, J.; Franci, C.; Huang, B.; Huang, J.; Yam, G.C.; et al. Critical Role of the Ubiquitin Ligase Activity of UHRF1, a Nuclear RING Finger Protein, in Tumor Cell Growth. Mol. Biol. Cell 2005, 16, 5621–5629.
- Du, Z.; Song, J.; Wang, Y.; Zhao, Y.; Guda, K.; Yang, S.; Kao, H.-Y.; Xu, Y.; Willis, J.; Markowitz, S.D.; et al. DNMT1 Stability Is Regulated by Proteins Coordinating Deubiquitination and Acetylation-Driven Ubiquitination. Sci. Signal. 2010, 3, ra80.
- Bostick, M.; Kim, J.K.; Estève, P.-O.; Clark, A.; Pradhan, S.; Jacobsen, S.E. UHRF1 Plays a Role in Maintaining DNA Methylation in Mammalian Cells. Science 2007, 317, 1760–1764.
- Sharif, J.; Muto, M.; Takebayashi, S.-I.; Suetake, I.; Iwamatsu, A.; Endo, T.A.; Shinga, J.; Mizutani-Koseki, Y.; Toyoda, T.; Okamura, K.; et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 2007, 450, 908–912.
- Karagianni, P.; Amazit, L.; Qin, J.; Wong, J. ICBP90, a Novel Methyl K9 H3 Binding Protein Linking Protein Ubiquitination with Heterochromatin Formation. Mol. Cell. Biol. 2008, 28, 705–717.
- Rothbart, S.B.; Krajewski, K.; Nady, N.; Tempel, W.; Xue, S.; Badeaux, A.I.; Barsyte-Lovejoy, D.; Martinez, J.Y.; Bedford, M.T.; Fuchs, S.M.; et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat. Struct. Mol. Biol. 2012, 19, 1155–1160.
- Rottach, A.; Frauer, C.; Pichler, G.; Bonapace, I.M.; Spada, F.; Leonhardt, H. The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res. 2009, 38, 1796–1804.
- Gelato, K.A.; Tauber, M.; Ong, M.S.; Winter, S.; Hiragami-Hamada, K.; Sindlinger, J.; Lemak, A.; Bultsma, Y.; Houliston, S.; Schwarzer, D.; et al. Accessibility of Different Histone H3-Binding Domains of UHRF1 Is Allosterically Regulated by Phosphatidylinositol 5-Phosphate. Mol. Cell 2014, 54, 905–919.
- Mistry, H.; Tamblyn, L.; Butt, H.; Sisgoreo, D.; Gracias, A.; Larin, M.; Gopalakrishnan, K.; Hande, M.P.; McPherson, J.P. UHRF1 is a genome caretaker that facilitates the DNA damage response to γ-irradiation. Genome Integr. 2010, 1, 7.
- Yang, C.; Wang, Y.; Zhang, F.; Sun, G.; Li, C.; Jing, S.; Liu, Q.; Cheng, Y. Inhibiting UHRF1 expression enhances radiosensitivity in human esophageal squamous cell carcinoma. Mol. Biol. Rep. 2013, 40, 5225–5235.
- Tian, Y.; Paramasivam, M.; Ghosal, G.; Chen, D.; Shen, X.; Huang, Y.; Akhter, S.; Legerski, R.; Chen, J.; Seidman, M.M.; et al. UHRF1 Contributes to DNA Damage Repair as a Lesion Recognition Factor and Nuclease Scaffold. Cell Rep. 2015, 10, 1957–1966.
- Liang, C.-C.; Zhan, B.; Yoshikawa, Y.; Haas, W.; Gygi, S.P.; Cohn, M.A. UHRF1 Is a Sensor for DNA Interstrand Crosslinks and Recruits FANCD2 to Initiate the Fanconi Anemia Pathway. Cell Rep. 2015, 10, 1947–1956.
- Motnenko, A.; Liang, C.-C.; Yang, D.; Lopez-Martinez, D.; Yoshikawa, Y.; Zhan, B.; Ward, K.E.; Tian, J.; Haas, W.; Spingardi, P.; et al. Identification of UHRF2 as a novel DNA interstrand crosslink sensor protein. PLoS Genet. 2018, 14, e1007643.
- Zhang, H.; Liu, H.; Chen, Y.; Yang, X.; Wang, P.; Liu, T.; Deng, M.; Qin, B.; Correia, C.; Lee, S.; et al. A cell cycle-dependent BRCA1–UHRF1 cascade regulates DNA double-strand break repair pathway choice. Nat. Commun. 2016, 7, 10201.
- Hahm, J.Y.; Kim, J.-Y.; Park, J.W.; Kang, J.-Y.; Kim, K.-B.; Kim, S.-R.; Cho, H.; Seo, S.-B. Methylation of UHRF1 by SET7 is essential for DNA double-strand break repair. Nucleic Acids Res. 2019, 47, 184–196.
- Hahm, J.Y.; Kang, J.-Y.; Park, J.W.; Jung, H.; Seo, S.-B. Methylated-UHRF1 and PARP1 Interaction Is Critical for Homologous Recombination. BMB Rep. 2020, 53, 112–117.
