Basophils and mast cells are among the principal inducers of Th2 responses and have a crucial role in allergic and anti-parasitic protective immunity. Basophils can function as antigen-presenting cells that bind antigens on their surface and boost humoral immune responses, inducing Th2 cell differentiation. Their depletion results in lower humoral memory activation and greater infection susceptibility. Basophils seem to have an active role upon immune response to SARS-CoV-2. In fact, a coordinate adaptive immune response to SARS-CoV-2 is magnified by basophils. It has been observed that basophil amount is lower during acute disease with respect to the recovery phase and that the grade of this depletion is an important determinant of the antibody response to the virus. Moreover, mast cells, present in a great quantity in the nasal epithelial and lung cells, participate in the first immune response to SARS-CoV-2. Their activation results in a hyperinflammatory syndrome through the release of inflammatory molecules, participating to the “cytokine storm” and, in a longer period, inducing pulmonary fibrosis.
- basophils
- mast cells
- COVID-19
- innate immune response
- adaptive immune response
1. Introduction
2. Basophils and COVID-19 Disease
Indeed, according to previous studies on viral infections, basophil depletion might impair the efficacy of IgG-responses to SARS-CoV-2. Considering a comprehensive point of view, basophils could enhance a coordinate adaptive immune response to SARS-CoV-2 that could be suppressed by the hyperinflammatory reaction during the acute phase of COVID-19. Further investigation will be required to understand the mechanisms of basophils in modulating humoral responses to SARS-CoV-2. Interestingly, as the expression of these cells seems to be lower during acute disease, it could be worth determining the grade of this depletion as an important determinant of the antibody response to the virus [42][18]. In the study from Wuhan No. 1 Hospital [43][19], involving 59 male and 68 female Chinese patients, a reduction in basophil count was present in 13.39% of enrolled patients. Moreover, authors noted that the reduction in basophils was present in the first three days of hospitalization and was restored to normal shortly. Fátima Conceição-Silva et al. [44][20] have shown that neutrophils, macrophages, lymphocytes, eosinophils, basophils, and mast cells can produce extracellular traps (ET), even if the modalities are still not completely known. Patients with severe cases of COVID-19 are predisposed to thrombosis in which ETs produced by neutrophils may participate. Contrary to neutrophils, ETs produced by basophils have a protective role against some infections with bactericidal and antifungal activity. A similar activity can also be hypothesized during COVID-19. A multidimensional analysis performed on laboratory parameters and diagnostic test of 178.887 Brazilian individuals, of whom 33.266 resulted in being positive for SARS-CoV 2 [45][21]. A case–control study [46][22] involving 74 COVID-19 patients and 228 non-COVID-19 patients, showed a significant reduction of basophil levels in COVID-19 patients, a result more evident in men older than 25 years of age in the Brazilian study. A similar result was also found in a retrospective study on 120 COVID-19 patients, 100 influenza patients, and 61 healthy controls: basophils were lower both in the COVID-19 group and in the influenza group as compared to controls [47][23]. To further support these findings, researchers conducted an observational, multicentric study [48][24] that compared levels of complete blood count and granulocytes subsets with cytofluorimetric analysis in COVID-19 patients and healthy blood donors as controls. The authors showed a significant decrease in basophil levels in COVID-19 patients when compared to controls. Similar findings came from a retrospective study [49][25] on 548 patients diagnosed with COVID-19 disease performed by Chen et al., in which a difference between on admission and end-hospitalization levels of basophils greater than 0.02 × 109/L (HR, 2.73; 95% CI, 1.5–6.47) represented a risk factor for fatal outcome, thus suggesting that the less is the basophil count on admission the poorer is the outcome of the patient. Analyzing severe cases of COVID-19 disease, in contrast with other low respiratory tract infections and excluding potentially confounding factors such as atopy and use of antihistamine drugs, Laing et al. [50][26] showed a dramatic depletion of plasmacytoid dendritic cells and basophils. A similar lower percentage of basophils in the white blood cell count was found by Qin et al. [51][27] in an observational study involving 452 patients in severe patients compared to non-severe cases (0.1 vs. 0.2%; p = 0.015). Further strength to these results is carried by the study of Sun Y. et al. [52][28] that evaluated the causal association between the different white blood cells and the COVID-19 susceptibility and severity by performing two-sample bidirectional Mendelian randomization analyses from the largest and most recent genome-wide association studies. Considering both severe COVID-19 disease and hospitalization due to COVID-19 disease as outcomes, the authors found an inverse association with low basophil count and low basophil percentage on white blood cell count ((OR = 0.75, CI: 0.60–0.95, p = 0.015), (OR = 0.70, CI: 0.54–0.92, p = 0.011)) and ((OR = 0.83, CI: 0.71–0.97, p = 0.020), (OR = 0.78, CI: 0.65–0.93, p = 0.005), respectively). The authors suggested a possible causal role of the reduced basophil count in increasing the risk of severe COVID-19 disease, potentially due to an insufficient innate immune response to SARS-CoV-2. No associations were found with COVID-19 susceptibility among white blood cells. Contrary to what was observed for basophils, an histopathological study conducted on 6 SARS-CoV-2-confirmed patients compared to 10 H1N1-infected patients and a control group of 10 patients who died for neoplastic or cardiovascular diseases [53][29], showed a striking increase in the number of mast cells in lung of COVID-19 patients. Specifically, mast cells, even in the degranulated form, were more frequently localized in the perivascular spaces between the alveolar sacs and terminal bronchioles and in the alveolar septa, close to the alveolar capillaries. Authors suggested that mast cells may play an important role in triggering the systemic cytokine storm associated with severe COVID-19 because of their production of various mediators, including IL-4 and IL-6, two cytokines involved in COVID-19 disease pathogenesis [54][30]. As previously described, when SARS-CoV-2 infects the host, the host is firstly attacked by innate immune cells, including mast cells. The latter are well expressed by nasal epithelial and lung cells and their activation may be responsible of hyperinflammatory syndrome [55,56][31][32]. Mast cells are ubiquitous in the body, and they are involved in several conditions including viral infections, systemic inflammatory diseases, asthma, neuroinflammatory diseases, traumatic brain injury, stroke, and several stress disorders [44,56][20][32]. Hence, these cells proved to be very heterogeneous, as they may differ in ultrastructure, morphology, mediator content, receptor expression, and responses to various stimuli, thus explaining the different effect they may cause, protective or damaging. In fact, mast cell cytoplasm is enriched of histamine, proteases, heparin, chondroitin sulfate, and proinflammatory and anti-inflammatory cytokines/chemokines filled into granules or de novo produced upon activation, which are released in response to stimuli [57,58,59][33][34][35]. While physiologically these molecules are useful to defeat viral and microbial threats, they can also mediate the inflammatory damage typical of asthma and allergic reactions. Through the same mechanism, SARS-CoV-2 may exploit the same effect of mast cells, inducing an uncontrolled inflammation activation mediated by the release of histamines, proteases, cytokines, chemokines, and arachidonic acid compounds such as prostaglandin D2 and leukotrienes [60,61,62][36][37][38]. Indeed, current reports highlighted that COVID-19 can activate mast cells through TLRs and contribute to pulmonary inflammation and fibrosis [63,64][39][40]. It also emerged, both in basophils and eosinophils, a reduction in the expression of surface CRTH2, a receptor for prostaglandin D2, and a significant increase in programmed cell death ligand 1 (PD-L1) in the severe form of the disease in contrast with mild group. Interestingly, both WHO and SOFA scores correlated with these two findings: positively with PD-L1 and negatively with CRTH2, respectively (Figure 1). The authors suggested that one possible mechanism, shared by other viruses as an escape mechanism, may be the increase of immune checkpoint levels, which usually prevent immune-driven diseases after immune response in tissue in order to avoid the clearance of viral particles during the infection [65,66][41][42]. Moreover, the downregulation of basophil and eosinophil CRTH2, an important activator of T helper 2 polarized response, induces speculation about a possible inhibition of this cell subset by SARS-CoV-2. The authors concluded that this latter mechanism, which is present in allergy and hyper-eosinophilic asthma, can possibly be responsible for the protective effect conferred against SARS-CoV-2 infection and severity by these conditions [67,68][43][44].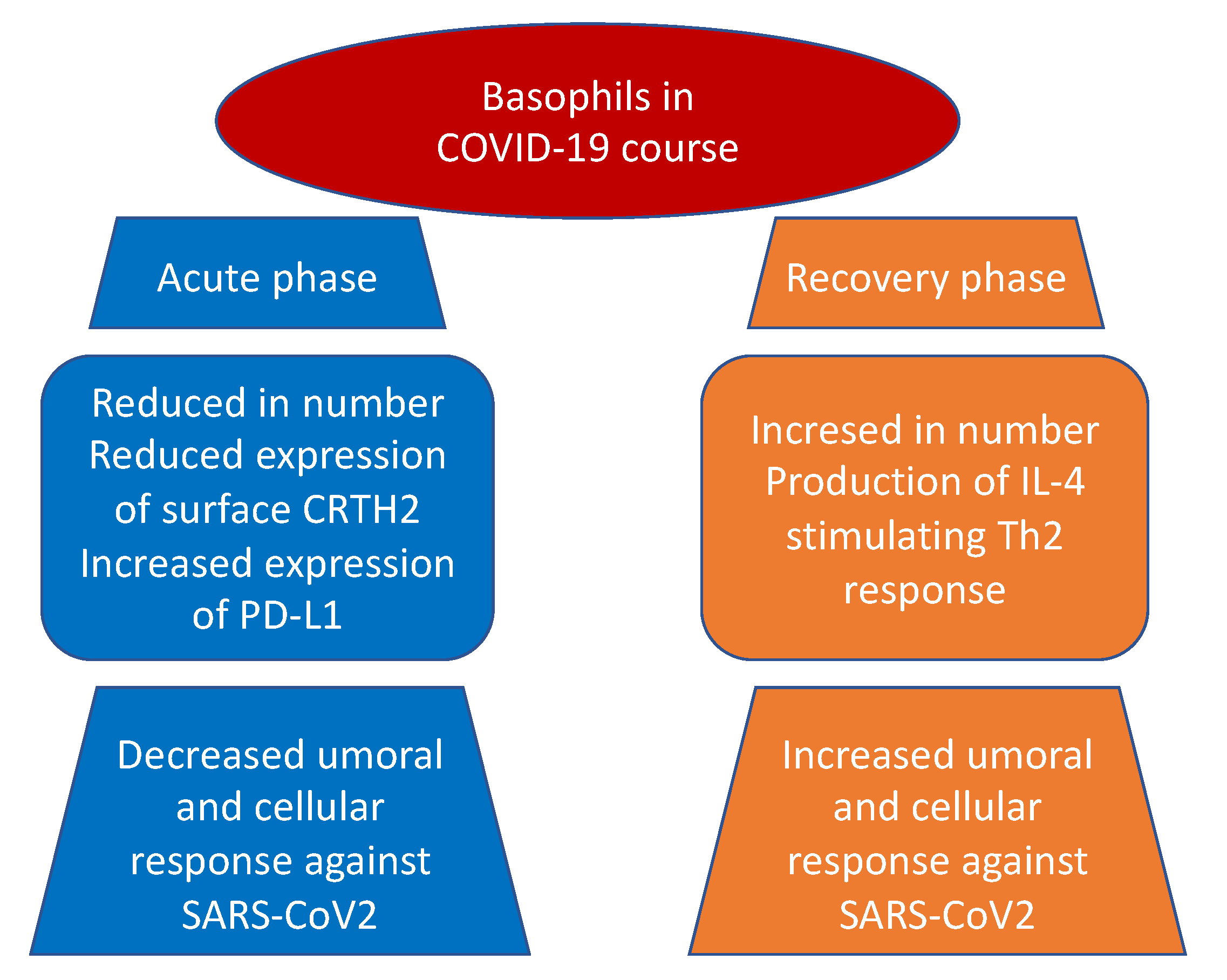
3. Mast Cell and COVID-19 Disease
The local inflammatory response in the lung observed in SARS-CoV-2-infected patients is characterized by a complex network of activated inflammatory innate immune cells, fibroblasts, endothelial cells, and bronchial epithelial cells. Bronchial epithelial cells and fibroblasts activated by SARS-CoV-2 cause upregulation of pro-inflammatory cytokines and induction of differentiation of mast cells that release histamine, proteases, cytokines, chemokines, and arachidonic acid compounds, such as prostaglandin D2 and leukotrienes, all of which are involved in the inflammatory network. Histamine is and is released into the vessels after cell stimulation. Histamine, stored endogenously within the secretory granules of mast cells, is involved in the increased expression of chemokine IL-8 and cytokine IL-6, thus favoring the hyperinflammation in the lung. Therefore, in the context of COVID-19 cytokine storm and severe disease, mast cells may act negative factors (Figure 2) as productors of histamine that induce microvascular leakage, proteases, and IL-6 that can degrade matrix, thus favoring intra-alveolar formation of the hyaline membrane and perpetuating inflammation, angiogenic factors and pro-coagulative factors, respectively, that may trigger immune thrombosis [75,76][53][54].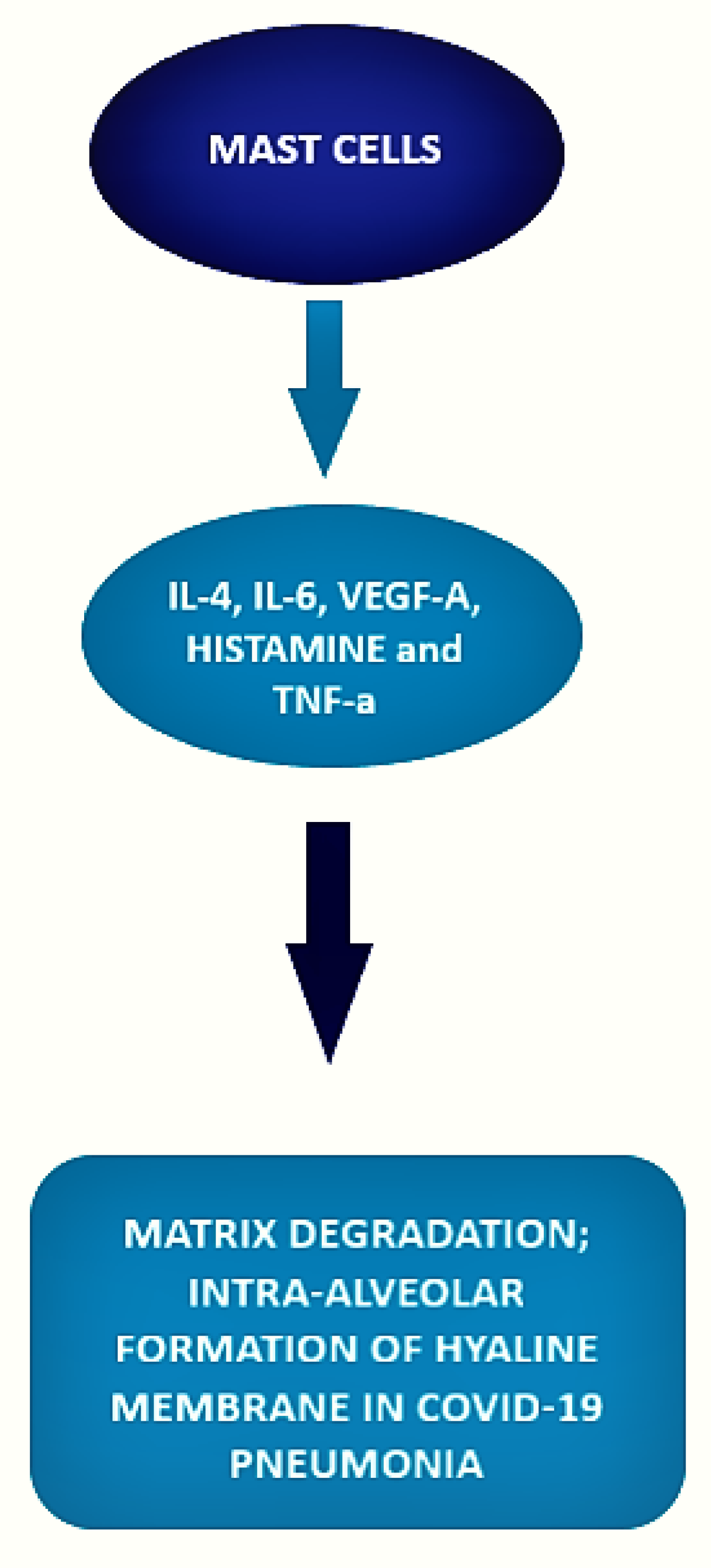
References
- Phua, J.; Weng, L.; Ling, L.; Egi, M.; Lim, C.-M.; Divatia, J.V.; Shrestha, B.R.; Arabi, Y.M.; Ng, J.; Gomersall, C.D.; et al. Intensive care management of coronavirus disease 2019 (COVID-19): Challenges and recommendations. Lancet Respir. Med. 2020, 8, 506–517.
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506.
- Chung, J.Y.; Thone, M.N.; Kwon, Y.J. COVID-19 vaccines: The status and perspectives in delivery points of view. Adv. Drug Deliv. Rev. 2020, 170, 1–25.
- Marovich, M.; Mascola, J.R.; Cohen, M.S. Monoclonal Antibodies for Prevention and Treatment of COVID-19. JAMA 2020, 324, 131–132.
- Tregoning, J.S.; Brown, E.S.; Cheeseman, H.M.; Flight, K.E.; Higham, S.L.; Lemm, N.M.; Pierce, B.F.; Stirling, D.C.; Wang, Z.; Pollock, K.M. Vaccines for COVID-19. Clin. Exp. Immunol. 2020, 202, 162–192.
- Brant, A.C.; Tian, W.; Majerciak, V.; Yang, W.; Zheng, Z.M. SARS-CoV-2: From its discovery to genome structure, transcrip-tion, and replication. Cell Biosci. 2021, 11, 136.
- Woo, P.C.; Lau, S.K.; Lam, C.S.; Lau, C.C.; Tsang, A.K.; Lau, J.H.; Bai, R.; Teng, J.L.; Tsang, C.C.; Wanget, M.; et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and be-tacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012, 86, 3995–4008.
- Mohamadian, M.; Chiti, H.; Shoghli, A.; Biglari, S.; Parsamanesh, N.; Esmaeilzadeh, A. COVID-19: Virology, biology and novel laboratory diagnosis. J. Gene Med. 2020, 23, e3303.
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220.
- Wang, M.-Y.; Zhao, R.; Gao, L.-J.; Gao, X.-F.; Wang, D.-P.; Cao, J.-M. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front. Cell. Infect. Microbiol. 2020, 10, 587269.
- Wiersinga, W.J.; Rhodes, A.; Cheng, A.C.; Peacock, S.J.; Prescott, H.C. Pathophysiology, Transmission, Diagnosis, and Treat-ment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020, 324, 782–793.
- Taghiloo, S.; Aliyali, M.; Abedi, S.; Mehravaran, H.; Sharifpour, A.; Zaboli, E.; Eslami-Jouybari, M.; Ghasemian, R.; Vahedi-Larijani, L.; Hossein-Nattaj, H.; et al. Apoptosis and immunophenotyping of peripheral blood lymphocytes in Iranian COVID-19 patients: Clinical and laboratory characteristics. J. Med. Virol. 2020, 93, 1589–1598.
- Zhang, H.; Wu, T. CD4+T, CD8+T counts and severe COVID-19: A meta-analysis. J. Infect. 2020, 81, e82–e84.
- Mazzoni, A.; Salvati, L.; Maggi, L.; Capone, M.; Vanni, A.; Spinicci, M.; Mencarini, J.; Caporale, R.; Peruzzi, B.; Antonelli, A.; et al. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J. Clin. Investig. 2020, 130, 4694–4703.
- Ragab, D.; Eldin, H.S.; Taeimah, M.; Khattab, R.; Salem, R. The COVID-19 Cytokine Storm; What We Know So Far. Front. Immunol. 2020, 11, 1446.
- Hu, B.; Huang, S.; Yin, L. The cytokine storm and COVID-19. J. Med. Virol. 2021, 93, 250–256.
- Allegra, A.; Di Gioacchino, M.; Tonacci, A.; Musolino, C.; Gangemi, S. Immunopathology of SARS-CoV-2 Infection: Immune Cells and Mediators, Prognostic Factors, and Immune-Therapeutic Implications. Int. J. Mol. Sci. 2020, 21, 13.
- Rodriguez, L.; Pekkarinen, P.T.; Lakshmikanth, T.; Tan, Z.; Consiglio, C.R.; Pou, C.; Chen, Y.; Mugabo, C.H.; Nguyen, N.A.; Nowlan, K.; et al. Systems-Level Immunomonitoring from Acute to Recovery Phase of Severe COVID-19. Cell Rep. Med. 2020, 1, 100078.
- Mao, J.; Dai, R.; Du, R.-C.; Zhu, Y.; Shui, L.-P.; Luo, X.-H. Hematologic changes predict clinical outcome in recovered patients with COVID-19. Ann. Hematol. 2021, 100, 675–689.
- Conceição-Silva, F.; Reis, C.; De Luca, P.; Leite-Silva, J.; Santiago, M.; Morrot, A.; Morgado, F. The Immune System Throws Its Traps: Cells and Their Extracellular Traps in Disease and Protection. Cells 2021, 10, 1891.
- Ten-Caten, F.; Gonzalez-Dias, P.; Castro, L.; Ogava, R.L.; Giddaluru, J.; Silva, J.C.S.; Martins, F.; Gonçalves, A.N.; Costa-Martins, A.G.; Araujo, J.D.; et al. In-depth analysis of laboratory parameters reveals the interplay between sex, age, and systemic inflammation in individuals with COVID-19. Int. J. Infect. Dis. 2021, 105, 579–587.
- Alnor, A.; Sandberg, M.B.; Toftanes, B.E.; Vinholt, P.J. Platelet parameters and leukocyte morphology is altered in COVID-19 patients compared to non-COVID-19 patients with similar symptomatology. Scand. J. Clin. Lab. Investig. 2021, 81, 213–217.
- Kazancioglu, S.; Bastug, A.; Ozbay, B.O.; Kemirtlek, N.; Bodur, H. The role of haematological parameters in patients with COVID-19 and influenza virus infection. Epidemiology Infect. 2020, 148, 1–16.
- Vitte, J.; Diallo, A.B.; Boumaza, A.; Lopez, A.; Michel, M.; Allardet-Servent, J.; Mezouar, S.; Sereme, Y.; Busnel, J.-M.; Miloud, T.; et al. A Granulocytic Signature Identifies COVID-19 and Its Severity. J. Infect. Dis. 2020, 222.
- Chen, R.; Sang, L.; Jiang, M.; Yang, Z.; Jia, N.; Fu, W.; Xie, J.; Guan, W.; Liang, W.; Ni, Z.; et al. Longitudinal hematologic and immunologic variations associated with the progression of COVID-19 patients in China. J. Allergy Clin. Immunol. 2020, 146, 89–100.
- Laing, A.G.; Lorenc, A.; del Molino Del Barrio, I.; Das, A.; Fish, M.; Monin, L.; Muñoz-Ruiz, M.; McKenzie, D.R.; Hayday, T.S.; Francos-Quijorna, I.; et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020, 26, 1623–1635.
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020, 71, 762–768.
- Sun, Y.; Zhou, J.; Ye, K. White Blood Cells and Severe COVID-19: A Mendelian Randomization Study. J. Pers. Med. 2021, 11, 195.
- Junior, J.D.S.M.; Miggiolaro, A.F.R.D.S.; Nagashima, S.; De Paula, C.B.V.; Baena, C.P.; Scharfstein, J.; De Noronha, L. Mast Cells in Alveolar Septa of COVID-19 Patients: A Pathogenic Pathway That May Link Interstitial Edema to Immunothrombosis. Front. Immunol. 2020, 11, 2369.
- Wang, J.; Lindholt, J.S.; Sukhova, G.K.; Shi, M.A.; Xia, M.; Chen, H.; Xiang, M.; He, A.; Wang, Y.; Xiong, N.; et al. IgE actions on CD4+ T cells, mast cells, and mac-rophages participate in the pathogenesis of experimental abdominal aortic aneurysms. EMBO Mol. Med. 2014, 6, 952–969.
- Kempuraj, D.; Selvakumar, G.P.; Ahmed, M.E.; Raikwar, S.P.; Thangavel, R.; Khan, A.; Zaheer, S.A.; Iyer, S.S.; Burton, C.; James, D.; et al. COVID-19, Mast Cells, Cyto-kine Storm, Psychological Stress, and Neuroinflammation. Neuroscientist 2020, 26, 402–414.
- Arac, A.; Grimbaldeston, M.A.; Galli, S.J.; Bliss, T.M.; Steinberg, G.K. Meningeal Mast Cells as Key Effectors of Stroke Pathol-ogy. Front. Cell Neurosci. 2019, 13, 126.
- Varricchi, G.; Rossi, F.W.; Galdiero, M.R.; Granata, F.; Criscuolo, G.; Spadaro, G.; de Paulis, A.; Marone, G. Physiological Roles of Mast Cells: Col-legium Internationale Allergologicum Update 2019. Int. Arch. Allergy Immunol. 2019, 179, 247–261.
- Gilfillan, A.M.; Austin, S.J.; Metcalfe, D.D. Mast Cell Biology: Introduction and Overview. Mast Cell Biol. 2011, 716, 2–12.
- Okayama, Y.; Kawakami, T. Development, Migration, and Survival of Mast Cells. Immunol. Res. 2006, 34, 97–116.
- Conti, P.; Caraffa, A.; Tetè, G.; Gallenga, C.E.; Ross, R.; Kritas, S.K.; Frydas, I.; Younes, A.; Di Emidio, P.; Ronconi, G. Mast cells activated by SARS-CoV-2 release hista-mine which increases IL-1 levels causing cytokine storm and inflammatory reaction in COVID-19. J. Biol. Regul. Homeost. Agents 2020, 34, 1629–1632.
- Sun, X.; Wang, T.; Cai, D.; Hu, Z.; Chen, J.; Liao, H.; Zhi, L.; Wei, H.; Zhang, Z.; Qiu, Y.; et al. Cytokine storm intervention in the early stages of COVID-19 pneumonia. Cytokine Growth Factor Rev. 2020, 53, 38–42.
- Theoharides, T.C. COVID -19, pulmonary mast cells, cytokine storms, and beneficial actions of luteolin. BioFactors 2020, 46, 306–308.
- Gigante, A.; Aquili, A.; Farinelli, L.; Caraffa, A.; Ronconi, G.; Gallenga, E.C.; Tetè, G.; Kritas, S.K.; Conti, P. Sodium chromo-glycate and palmitoyleth-anolamide: A possible strategy to treat mast cell-induced lung inflammation in COVID-19. Med. Hypotheses 2020, 143, 109856.
- Karamloo, F.; Konig, R. SARS-CoV-2 immunogenicity at the crossroads. Allergy 2020, 75, 1822–1824.
- Wang, Z.; Wang, S.; Goplen, N.P.; Li, C.; Cheon, I.S.; Dai, Q.; Huang, S.; Shan, J.; Ma, C.; Ye, Z.; et al. PD-1(hi) CD8(+) resident memory T cells balance immuni-ty and fibrotic sequelae. Sci Immunol. 2019, 4, 36.
- Kahan, S.M.; Zajac, A.J. Immune Exhaustion: Past Lessons and New Insights from Lymphocytic Choriomeningitis Virus. Viruses 2019, 11, 156.
- Jackson, D.J.; Busse, W.W.; Bacharier, L.B.; Kattan, M.; O’Connor, G.T.; Wood, R.A.; Visness, C.M.; Durham, S.R.; Larson, D.; Esnault, S.; et al. Association of respiratory allergy, asthma, and expression of the SARS-CoV-2 receptor ACE2. J. Allergy Clin. Immunol. 2020, 146, 203–206.e3.
- Carli, G.; Cecchi, L.; Stebbing, J.; Parronchi, P.; Farsi, A. Is asthma protective against COVID-19? Allergy 2021, 76, 866–868.
- Denzel, A.; Maus, U.A.; Gomez, M.R.; Moll, C.; Niedermeier, M.; Winter, C.; Maus, R.; Hollingshead, S.; Briles, D.E.; Kunz-Schughart, L.A.; et al. Basophils enhance immunological memory responses. Nat. Immunol. 2008, 9, 733–742.
- Kawakami, T. Basophils now enhance memory. Nat. Immunol. 2008, 9, 720–721.
- Dufour, J.H.; Dziejman, M.; Liu, M.T.; Leung, J.H.; Lane, T.E.; Luster, A.D. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J. Immunol. 2002, 168, 3195–3204.
- Angiolillo, A.L.; Sgadari, C.; Taub, D.D.; Liao, F.; Farber, J.M.; Maheshwari, S.; Kleinman, H.K.; Reaman, G.H.; Tosato, G. Human interferon-inducible protein 10 is a potent inhibitor of angiogenesis in vivo. J. Exp. Med. 1995, 182, 155–162.
- Carsana, L.; Sonzogni, A.; Nasr, A.; Rossi, R.S.; Pellegrinelli, A.; Zerbi, P.; Rech, R.; Colombo, R.; Antinori, S.; Corbellino, M.; et al. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: A two-centre descriptive study. Lancet Infect. Dis. 2020, 20, 1135–1140.
- Crivellato, E.; Travan, L.; Ribatti, D. Mast cells and basophils: A potential link in promoting angiogenesis during allergic in-flammation. Int. Arch. Allergy Immunol. 2010, 151, 89–97.
- Swystun, L.L.; Liaw, P.C. The role of leukocytes in thrombosis. Blood 2016, 128, 753–762.
- Merle, N.S.; Noé, R.; Halbwachs-Mecarelli, L.; Fremeaux-Bacchi, V.; Roumenina, L.T. Complement System Part II: Role in Immunity. Front. Immunol. 2015, 6, 257.
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological mis-firing in severe COVID-19. Nature 2020, 584, 463–469.
- Voehringer, D. Protective and pathological roles of mast cells and basophils. Nat. Rev. Immunol. 2013, 13, 362–375.
