Magnetic resonance imaging (MRI) is the most important paraclinical support in multiple sclerosis (MS) diagnostic work-up. Still, its specificity using conventional sequences is limited as the cerebral white matter (WM) findings in patients with different morbidities may be mistakenly interpreted as MS lesions. Therefore, more MS-specific MRI criteria to rule out disorders are needed. The central vein sign (CVS) has been proposed as an MS imaging biomarker to differentiate MS from MS-radiological mimicking disorders, implementing susceptibility-based MRI sequences. In this study, we aim to systematically review the proportion of MS lesions which show the CVS and estimate the performance diagnostic value in discriminating MS from its common radiological differential diagnosis. Also, we investigate the impact of using different magnetic field strengths, specific sequences and post-processing techniques.
- multiple sclerosis
- central vein sign
- imaging
- MRI
- biomarker
1. Introduction
Evaluating the role of more specific magnetic resonance imaging (MRI) features of multiple sclerosis (MS) lesions, such as the central vein sign (CVS), is considered a high-priority area for research according to The International Panel on Diagnosis of MS, which produced the 2017 revised diagnostic criteria[1].
The central vein sign is a marker developed initially in 7 Tesla (T) MRI studies[2][3], relying on the pathological specificity of perivenular distribution of MS lesions, exploiting the ability of susceptibility (T2*-“T2-star”)-based MRI sequences to highlight in vivo paramagnetic properties of deoxyhemoglobin in venous blood flow[4].
By combining both magnitude and phase images acquired using T2*-weighted gradient echo (GRE) pulse sequences, it is possible to exploit susceptibility-weighted imaging (SWI) that can reveal the presence of blood vessels in MS lesions[5][6]. Even though the core protocol always includes a GRE sequence, many MRI parameters can be utilized to obtain SWI[5]. For instance, some studies introduced echo-planar imaging (EPI) to retrieve high-resolution images while reducing the scan time[7]. The post-processing implementation introduces further heterogeneity in the studies. The first processing step required to compute SWI is the removal of artefacts from phase images. Several different phase processing approaches have been tested where high-pass filters are applied directly to phase images or following phase unwrapping [8]. Besides obtaining SWI from the combination of both magnitude and phase from the same GRE acquisition, another strategy consists of multiplying a T2*-weighted sequence with the fluid-attenuated inversion recovery (FLAIR) sequence to obtain a FLAIR* image[9]. This approach combines the sensibility of FLAIR in detecting WM lesions and the ability of susceptibility-based imaging to highlight the presence of the vein (Figure 1).
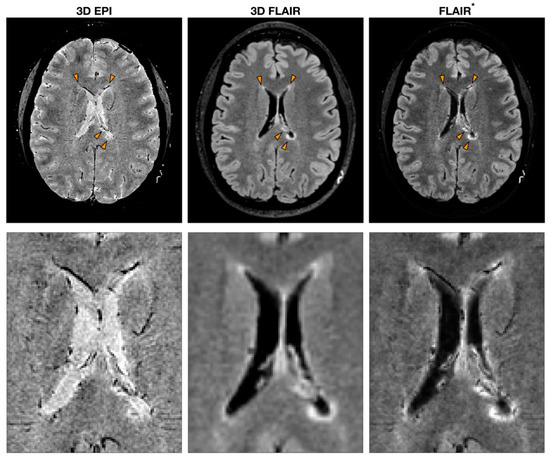
Figure 1.
Example of MS MRI images obtained with different sequences to detect the central vein sign (CVS). T2*-weighted 3D echo-planar imaging (EPI), 3D fluid-attenuated inversion recovery (FLAIR) and FLAIR*, a post-processing technique that combines the EPI sequence [or more generally a T2*w sequence (used to detect CVS)] and FLAIR. The orange arrowheads indicate MS lesions with the characteristic CVS. A zoomed-in version of the same image is reported to highlight the CVS feature for each modality.
Although a definition of the CVS has been recently provided[10], and criteria based on both proportion lesion threshold[11] [12] or lesion number threshold [13][14] have been proposed to discriminate MS from its MRI mimics, its use in clinical practice has yet to be implemented.
2. Discussion
The systematic review and meta-analysis (35 studies considered eligible up to 24 August 2020) we performed revealed a proportion of 73% of MS lesions showing a CVS. Moreover, the use of the CVS reported a notable diagnostic performance, providing a pooled specificity of 92% and sensitivity of 95%. The pooled diagnostic performances in differentiating MS from other diseases estimated with the area under the HSROC of 0.98 (95%CI, 0.96–0.99%) revealed an excellent performance in this task. The optimal cut-off value obtained from the single patient data pooled together was 40% with an excellent value of the accuracy calculated by the area under the ROC (0.946). This cut-off value confirms this proportion threshold-based criteria as an excellent value with a sensitivity of 90% and a specificity of 89%. A portion of the studies included in this review used the FLAIR* approach[9], but the analyses included in this study did not provide any difference in the proportion of detectable CVS-positive lesions when using this approach with respect to others. The limited number of studies employing this method available for this review might have influenced this result; however, this approach might be rarely accessible in a clinical setting since to produce FLAIR* images, processing steps such as coregistration and multiplication, often not implemented directly in MRI scanners, are necessary. Another interesting aspect highlighted by this examination is that both the choice of the sequence and the scanner field strength play an important role in identifying the CVS. As expected, the higher signal-to-noise ratio and contrast-to-noise ratio of 7T MRI scanners do contribute to delineate better the CVS (0.82); however, we found that the use of 3T scanner might be sufficient in this regard (0.74). In contrast, 1.5T scanners showed a statistically significant reduction in CSV detection (0.58). Among sequences, we found a statistically significant improvement of the CVS proportion detected in MS lesions using a 3D-EPI sequence versus all other sequences (0.82 vs 0.71) regardless of the magnetic field strength. However, this particular sequence is not routinely used, perhaps due to a standard product sequence's unavailability from some manufacturers.
3. Conclusion
Despite the high heterogeneity of the studies included in this meta-analysis, the CVS in differentiating MS lesions from other confounding diseases could be used. We highlight that the minimum scanner field strength needed to better exploit the CVS specificity is 3T, and the T2*-weighted 3D-EPI sequence could be suggested as a preferable image acquisition strategy.
References
- Alan J Thompson; Brenda L Banwell; Frederik Barkhof; William M Carroll; Timothy Coetzee; Giancarlo Comi; Jorge Correale; Franz Fazekas; Massimo Filippi; Mark S Freedman; et al.Kazuo FujiharaSteven L GalettaHans Peter HartungLudwig KapposFred D LublinRuth Ann MarrieAaron E MillerDavid H MillerXavier MontalbanEllen M MowryPer Soelberg SorensenMar TintoréAnthony L TraboulseeMaria TrojanoBernard M J UitdehaagSandra VukusicEmmanuelle WaubantBrian G WeinshenkerStephen C ReingoldJeffrey A Cohen Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. The Lancet Neurology 2018, 17, 162-173, 10.1016/s1474-4422(17)30470-2.
- Kathryn E. Hammond; Meredith Metcalf; Lucas Carvajal; Darin T. Okuda; Radhika Srinivasan; Dan Vigneron; Sarah J. Nelson; Daniel Pelletier; Quantitative in vivo magnetic resonance imaging of multiple sclerosis at 7 Tesla with sensitivity to iron. Annals of Neurology 2008, 64, 707-713, 10.1002/ana.21582.
- Emma C. Tallantyre; M. J. Brookes; J. E. Dixon; P. S. Morgan; N. Evangelou; P. G. Morris; DEMONSTRATING THE PERIVASCULAR DISTRIBUTION OF MS LESIONS IN VIVO WITH 7-TESLA MRI. Neurology 2008, 70, 2076-2078, 10.1212/01.wnl.0000313377.49555.2e.
- J R Reichenbach; R Venkatesan; D J Schillinger; D K Kido; E M Haacke; Small vessels in the human brain: MR venography with deoxyhemoglobin as an intrinsic contrast agent.. Radiology 1997, 204, 272-277, 10.1148/radiology.204.1.9205259.
- Ewart Mark Haacke; S. Mittal; Z. Wu; J. Neelavalli; Y.-C.N. Cheng; Susceptibility-Weighted Imaging: Technical Aspects and Clinical Applications, Part 1. American Journal of Neuroradiology 2008, 30, 19-30, 10.3174/ajnr.a1400.
- S. Mittal; Z. Wu; J. Neelavalli; E.M. Haacke; Susceptibility-Weighted Imaging: Technical Aspects and Clinical Applications, Part 2. American Journal of Neuroradiology 2009, 30, 232-252, 10.3174/ajnr.a1461.
- P. Sati; D. M. Thomasson; N. Li; D. L. Pham; N. M. Biassou; D. S. Reich; J. A. Butman; Rapid, high-resolution, whole-brain, susceptibility-based MRI of multiple sclerosis. Multiple Sclerosis Journal 2014, 20, 1464-1470, 10.1177/1352458514525868.
- Ningzhi Li; Wen-Tung Wang; Pascal Sati; Dzung L. Pham; John A. Butman Md; Quantitative assessment of susceptibility-weighted imaging processing methods. Journal of Magnetic Resonance Imaging 2013, 40, 1463-1473, 10.1002/jmri.24501.
- P. Sati; Ilena C. George; Colin D. Shea; María I. Gaitán; Daniel S. Reich; FLAIR*: A Combined MR Contrast Technique for Visualizing White Matter Lesions and Parenchymal Veins. Radiology 2012, 265, 926-932, 10.1148/radiol.12120208.
- Pascal Sati; on behalf of the NAIMS Cooperative; Jiwon Oh; R. Todd Constable; Nikos Evangelou; Charles R. G. Guttmann; Roland G. Henry; Eric C. Klawiter; Caterina Mainero; Luca Massacesi; et al.Henry McFarlandFlavia NelsonDaniel OntanedaAlexander RauscherWilliam D. RooneyNikos Evangelou Amal P. R. SamaraweeraRussell T. ShinoharaRaymond A. SobelAndrew J. SolomonCaterina Mainero Constantina A. TreabaJens WuerfelRobert ZivadinovNancy L. SicotteDaniel PelletierPascal Sati Henry McFarland Daniel S. Reich The central vein sign and its clinical evaluation for the diagnosis of multiple sclerosis: a consensus statement from the North American Imaging in Multiple Sclerosis Cooperative. Nature Reviews Neurology 2016, 12, 714-722, 10.1038/nrneurol.2016.166.
- Pietro Maggi; Martina Absinta Md; Matteo Grammatico; Luisa Vuolo; Giacomo Emmi; Giovanna Carlucci Md; Gregorio Spagni; Alessandro Barilaro Md; Anna Maria Repice; Lorenzo Emmi; et al.Domenico PriscoVittorio MartinelliRoberta ScottiNiloufar Sadeghi MdGaetano PerrottaPascal SatiDaniel S ReichDaniel S. Reich MdMassimo FilippiLuca Massacesi Central vein sign differentiates Multiple Sclerosis from central nervous system inflammatory vasculopathies. Annals of Neurology 2018, 83, 283-294, 10.1002/ana.25146.
- E. C. Tallantyre; J. E. Dixon; I. Donaldson; T. Owens; P. S. Morgan; P. G. Morris; Nikos Evangelou; Ultra-high-field imaging distinguishes MS lesions from asymptomatic white matter lesions. Neurology 2011, 76, 534-539, 10.1212/wnl.0b013e31820b7630.
- Niraj Mistry; Rasha Abdel-Fahim; Amal Samaraweera; Olivier Mougin; Emma Tallantyre; Christopher Tench; Tim Jaspan; Peter Morris; Paul S Morgan; Nikos Evangelou; et al. Imaging central veins in brain lesions with 3-T T2*-weighted magnetic resonance imaging differentiates multiple sclerosis from microangiopathic brain lesions. Multiple Sclerosis Journal 2016, 22, 1289-1296, 10.1177/1352458515616700.
- Andrew J. Solomon; Richard Watts; Daniel Ontaneda; Martina Absinta Md; Pascal Sati; Daniel S Reich; Diagnostic performance of central vein sign for multiple sclerosis with a simplified three-lesion algorithm. Multiple Sclerosis Journal 2017, 24, 750-757, 10.1177/1352458517726383.
