Several times during the past two decades, epidemic viral diseases created global challenges. Although many solutions have been proposed to deal with this tight spot, it is still believed that public vaccination represents the most effective strategy to handle it. So far, various kinds of vaccines including protein subunits, virus-like particles, inactivated, live attenuated, viral vectors, RNA, and DNA vaccines have been used in the prevention of COVID-19. Among the various categories of vaccines, peptide vaccines have created a new hope for quick and trustworthy access due to the development of proteomics equipment. This review specifically focuses on vaccines and peptide therapies in severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). We consider here the efficacy and safety of subunit and synthetic peptides vaccine in clinical trial phases. Furthermore, monoclonal antibodies with the ability to suppress the development of SARS-CoV-2, those candidates that have entered into clinical trials until March 2023, were selected and evaluated..
- SARS-CoV-2
- peptide vaccines
- COVID-19 vaccination
1. Introduction
2. Methodological Routes to Discover and Finalize a Peptide as a Protective Agent
The idea of using peptides to stimulate the immune system has a very long history. However, the entry of this concept into the field of modern medicine is due to the studies of William Bradley Coley (1862–1936). Studies show that the peptides as vaccine candidates against SARS-CoV-2 are generally synthetic [15][14]. It is expected that the sequence of a peptide vaccine (which is between 20 and 30 amino acids) must be immunogenic or able to disrupt a stage of the virus’s path of pathogenesis. When it comes to the synthesis of peptide vaccines, it is expected that their production process will be carried out with higher precision and reproducibility, which will guarantee the quality of vaccination, as well as the speed of their industrialization [16][15]. However, studies on some of these types of peptides show that due to the not enough strong immune response they create, the presence of adjuvants is always needed to obtain an adequate response [15][14]. Another challenge that has been observed is their vulnerability to proteases in the body’s environment. As a result, most peptide vaccines do not have much resistance against proteases. However, it is suggested that the challenge can be overcome by inserting the vaccines into liposomes, conjugation on the surface of other carrier proteins, or connecting them to the surfaces of nanoparticles [17][16]. In this section, according to Figure 1, the methodological ways by which peptide vaccines can be proposed are presented.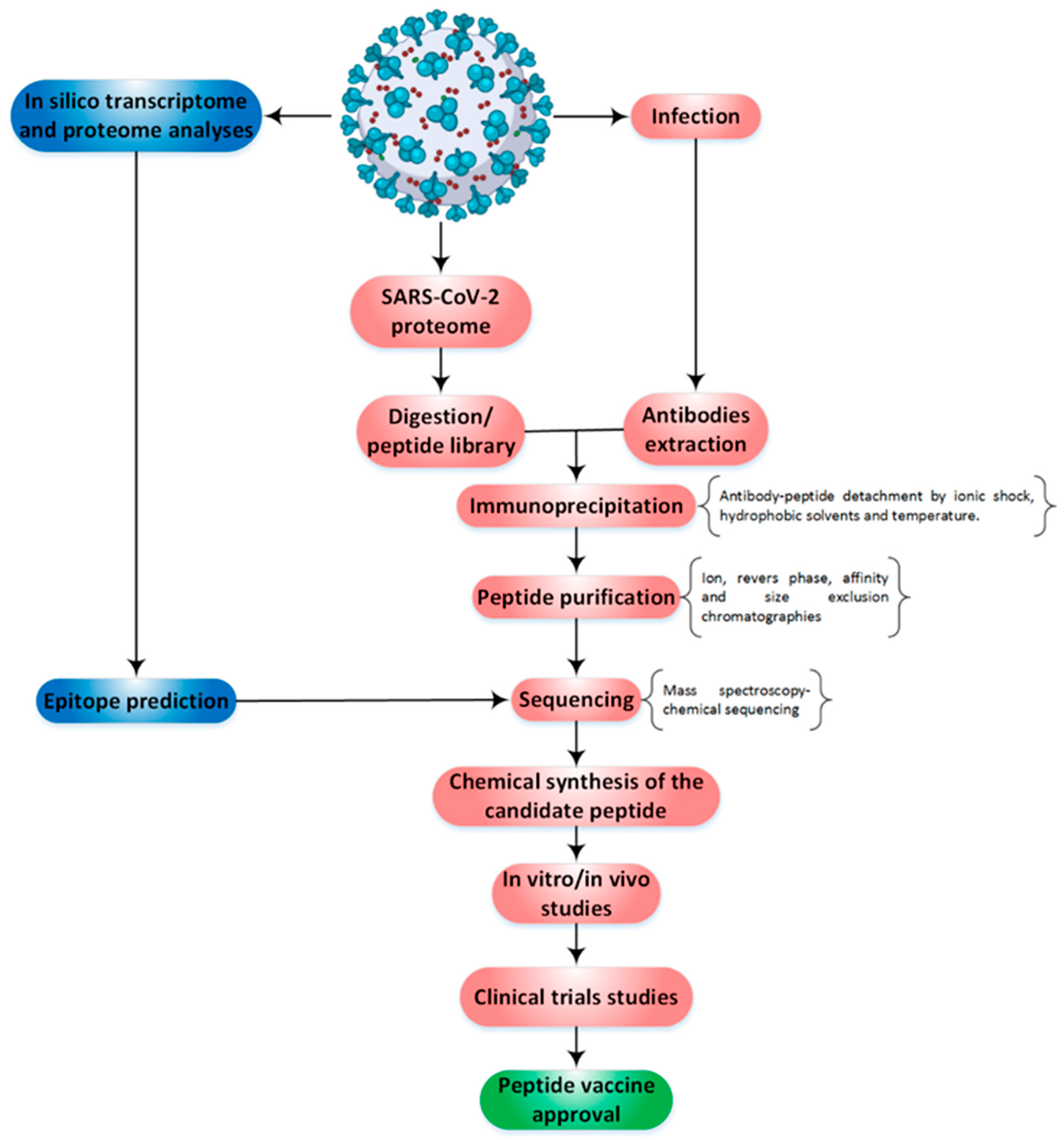
3. Landscape of SARS-CoV-2 Peptide Vaccines on Clinical Trials
Based on the document published by the World Health Organization (WHO) [24][22], by February 2022, 146 vaccines are being developed to the clinical stage, and 159 vaccines are currently under investigation in the pre-clinical phase [25,26][23][24]. The subunit protein platform comprises 33% of all vaccines in the clinical phase (see Figure 2).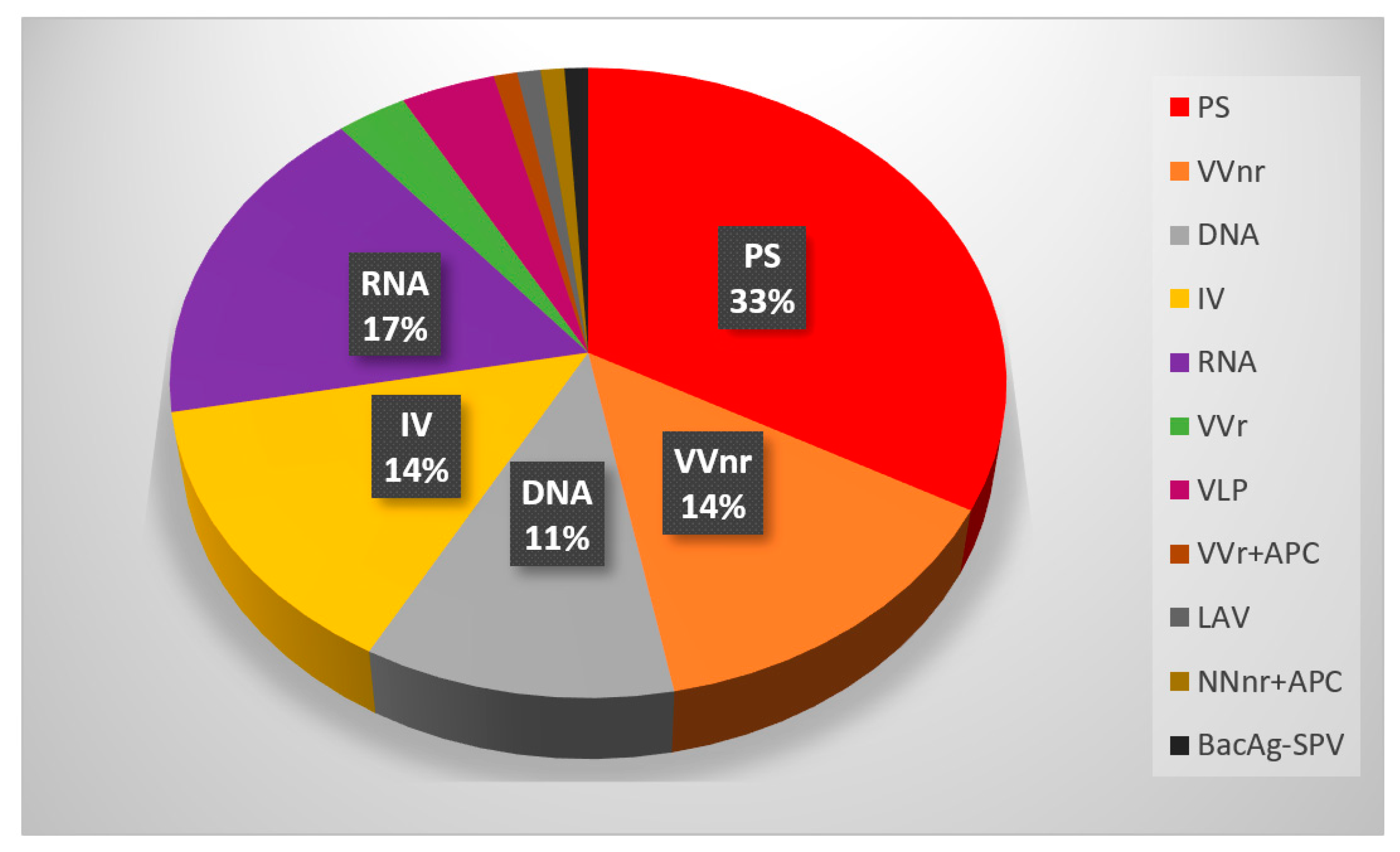
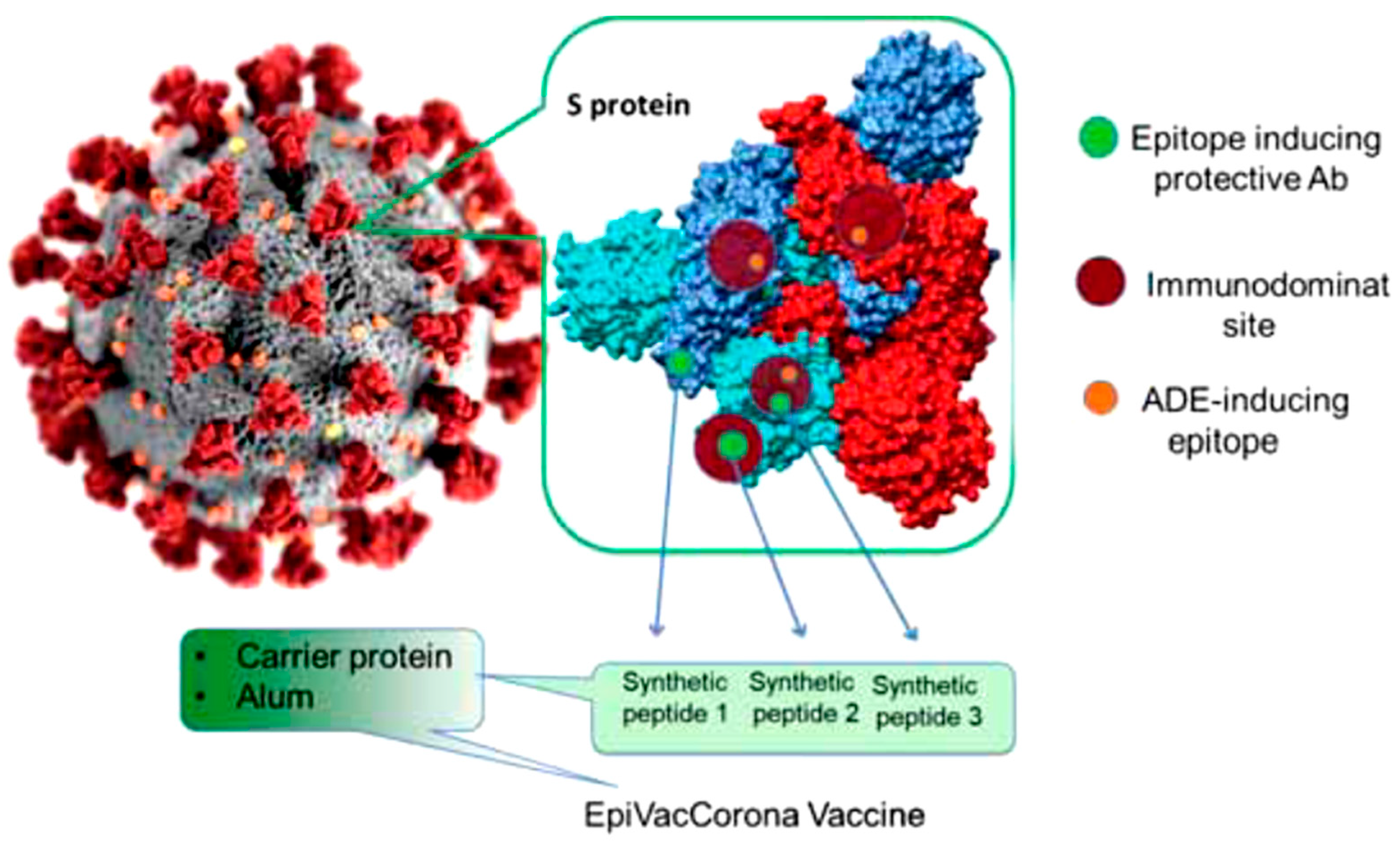
References
- Burrell, C.J.; Howard, C.R.; Murphy, F.A. Epidemiology of viral infections. Fenner White’s Med. Virol. 2017, 185–203.
- Zarkesh, K.; Entezar-Almahdi, E.; Ghasemiyeh, P.; Akbarian, M.; Bahmani, M.; Roudaki, S.; Fazlinejad, R.; Mohammadi-Samani, S.; Firouzabadi, N.; Hosseini, M.; et al. Drug-based therapeutic strategies for COVID-19-infected patients and their challenges. Future Microbiol. 2021, 16, 1415–1451.
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Coronaviruses Methods Protoc. 2015, 1–23.
- Zhang, S.F.; Tuo, J.L.; Huang, X.B.; Zhu, X.; Zhang, D.M.; Zhou, K.; Yuan, L.; Luo, H.J.; Zheng, B.J.; Yuen, K.Y.; et al. Epidemiology characteristics of human coronaviruses in patients with respiratory infection symptoms and phylogenetic analysis of HCoV-OC43 during 2010–2015 in Guangzhou. PLoS ONE 2018, 13, e0191789.
- Fung, T.S.; Liu, D.X. Human coronavirus: Host-pathogen interaction. Annu. Rev. Microbiol. 2019, 73, 529–557.
- Peiris, M.; Poon, L.L.M. Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) (Coronaviridae). Encycl. Virol. 2021, 814–824.
- World Health Organization. Coronavirus 2020. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 4 July 2020).
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733.
- Matricardi, P.M.; Dal Negro, R.W.; Nisini, R. The first, holistic immunological model of COVID-19: Implications for prevention, diagnosis, and public health measures. Pediatr. Allergy Immunol. 2020, 31, 454–470.
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224.
- Dong, M.; Zhang, J.; Ma, X.; Tan, J.; Chen, L.; Liu, S.; Xin, Y.; Zhuang, L. ACE2, TMPRSS2 distribution and extrapulmonary organ injury in patients with COVID-19. Biomed. Pharmacother. 2020, 131, 110678.
- Lucchese, G. Epitopes for a 2019-nCoV vaccine. Cell. Mol. Immunol. 2020, 17, 539–540.
- Song, Z.; Xu, Y.; Bao, L.; Zhang, L.; Yu, P.; Qu, Y.; Zhu, H.; Zhao, W.; Han, Y.; Qin, C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses 2019, 11, 59.
- Purcell, A.W.; McCluskey, J.; Rossjohn, J. More than one reason to rethink the use of peptides in vaccine design. Nat. Rev. Drug Discov. 2007, 6, 404–414.
- Nelde, A.; Rammensee, H.G.; Walz, J.S. The peptide vaccine of the future. Mol. Cell. Proteom. 2021, 20, 100022.
- Kianpour, M.; Akbarian, M.; Uversky, V.N. Nanoparticles for coronavirus control. Nanomaterials 2022, 12, 1602.
- Heitmann, J.S.; Bilich, T.; Tandler, C.; Nelde, A.; Maringer, Y.; Marconato, M.; Reusch, J.; Jäger, S.; Denk, M.; Richter, M.; et al. A COVID-19 peptide vaccine for the induction of SARS-CoV-2 T cell immunity. Nature 2022, 601, 617–622.
- Akbarian, M.; Lundstrom, K.; Redwan, E.M.; Uversky, V.N. Vaccine Development Strategies and the Current Status of COVID-19 Vaccines. In COVID-19: From Bench to Bedside; CRC Press: Boca Raton, FL, USA, 2022.
- Bartlam, M.; Xu, Y.; Rao, Z. Structural proteomics of the SARS coronavirus: A model response to emerging infectious diseases. J. Struct. Funct. Genom. 2007, 8, 85–97.
- Stukalov, A.; Girault, V.; Grass, V.; Bergant, V.; Karayel, O.; Urban, C.; Haas, D.A.; Huang, Y.; Oubraham, L.; Wang, A.; et al. Multi-level proteomics reveals host-perturbation strategies of SARS-CoV-2 and SARS-CoV. bioRxiv 2020, 1, 1.
- Dudek, N.L.; Croft, N.P.; Schittenhelm, R.B.; Ramarathinam, S.H.; Purcell, A.W. A systems approach to understand antigen presentation and the immune response. Proteom. Syst. Biol. Methods Protoc. 2016, 1394, 189–209.
- Worl Health Organization. Draft Landscape of COVID-19 Candidate Vaccines. 2 July 2020. Available online: https://www.who.int/publications/m/item/draft-landscape-of-COVID-19-candidate-vaccines (accessed on 4 July 2020).
- Fares, S.; Elmnyer, M.M.; Mohamed, S.S.; Elsayed, R. COVID-19 vaccination perception and attitude among healthcare workers in Egypt. J. Prim. Care Community Health 2021, 12, 21501327211013303.
- Yan, Z.P.; Yang, M.; Lai, C.L. COVID-19 vaccines: A review of the safety and efficacy of current clinical trials. Pharmaceuticals 2021, 14, 406.
- OSE Immunotherapeutics. A Randomized, Open Label, Phase 1 Study to Evaluate the Safety, Reactogenicity and Immunogenicity of OSE-13E, a Multiepitope-Based Vaccine Candidate against COVID-19, in Healthy Adults (COVEPIT-3). Clinical Trial Registration NCT04885361. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT04885361 (accessed on 26 June 2023).
- Belayachi, J.; Obtel, M.; Mhayi, A.; Razine, R.; Abouqal, R. Long term effectiveness of inactivated vaccine BBIBP-CorV (Vero Cells) against COVID-19 associated severe and critical hospitalization in Morocco. PLoS ONE 2022, 17, e0278546.
- Garnett, L.; Tran, K.; Chan, M.; Tierney, K.; Schiffman, Z.; Audet, J.; Lew, J.; Meilleur, C.; Chan, M.; Manguiat, K.; et al. An S1 subunit vaccine and combination adjuvant (COVAC-1) elicits robust protection against SARS-CoV-2 challenge in African green monkeys. bioRxiv 2022, 6.
- Luo, D.; Pan, H.; He, P.; Yang, X.; Li, T.; Ning, N.; Fang, X.; Yu, W.; Wei, M.; Gao, H. A randomized, double-blind, placebo-controlled phase 1 and phase 2 clinical trial to evaluate efficacy and safety of a SARS-CoV-2 vaccine SCoK in adults. Clin. Transl. Med. 2022, 12, e1016.
- Richmond, P.; Hatchuel, L.; Dong, M.; Ma, B.; Hu, B.; Smolenov, I.; Li, P.; Liang, P.; Han, H.H.; Liang, J.; et al. Safety and immunogenicity of S-Trimer (SCB-2019), a protein subunit vaccine candidate for COVID-19 in healthy adults: A phase 1, randomised, double-blind, placebo-controlled trial. Lancet 2021, 397, 682–694.
- A Controlled Phase 2/3 Study of Adjuvanted Recombinant SARS-CoV-2 Trimeric S-protein Vaccine (SCB-2019) for the Prevention of COVID-19. NCT04672395. Available online: www.ClinicalTrials.gov (accessed on 26 June 2023).
- Lopez, P.; Bravo, L.; Buntinx, E.; Borja-Tabora, C.; Velasquez, H.; Rodriguez, E.J.; Rodriguez, C.A.; Carlos, J.; Montellano, M.E.; Alberto, E.R. Safety and immunogenicity of SCB-2019, an adjuvanted, recombinant SARS-CoV-2 trimeric S-protein subunit COVID-19 vaccine in healthy 12–17 year-old adolescents. medRxiv 2023, 2, 23286317.
- Tabarsi, P.; Anjidani, N.; Shahpari, R.; Roshanzamir, K.; Fallah, N.; Andre, G.; Petrovsky, N.; Barati, S. Immunogenicity and safety of SpikoGen®, an adjuvanted recombinant SARS-CoV-2 spike protein vaccine as a homologous and heterologous booster vaccination: A randomized placebo-controlled trial. Immunology 2022, 167, 340–353.
- Tabarsi, P.; Anjidani, N.; Shahpari, R.; Mardani, M.; Sabzvari, A.; Yazdani, B.; Kafi, H.; Fallah, N.; Ebrahimi, A.; Taheri, A.; et al. Evaluating the efficacy and safety of SpikoGen®, an Advax-CpG55. 2–adjuvanted severe acute respiratory syndrome coronavirus 2 spike protein vaccine: A phase 3 randomized placebo-controlled trial. Clin. Microbiol. Infect. 2023, 29, 215–220.
- Mallory, R.M.; Formica, N.; Pfeiffer, S.; Wilkinson, B.; Marcheschi, A.; Albert, G.; McFall, H.; Robinson, M.; Plested, J.S.; Zhu, M.; et al. Safety and immunogenicity following a homologous booster dose of a SARS-CoV-2 recombinant spike protein vaccine (NVX-CoV2373): A secondary analysis of a randomised, placebo-controlled, phase 2 trial. Lancet Infect. Dis. 2022, 22, 1565–1576.
- Medicine UNLo. Evaluation of the Safety and Immunogenicity of a SARS-CoV-2 rS (COVID-19) Nanoparticle Vaccine with/without Matrix-M. Adjuvant. Available online: https://clinicaltrials.gov/ct2/show/NCT04368988 (accessed on 26 June 2023).
