Skin and wound infections are serious medical problems, and the diversity of bacteria makes such infections difficult to treat. Bacteria possess many virulence factors, among which motility plays a key role in skin infections. This feature allows for movement over the skin surface and relocation into the wound. The aim of this paper is to review the type of bacterial movement and to indicate the underlying mechanisms than can serve as a target for developing or modifying antibacterial therapies applied in wound infection treatment. Five types of bacterial movement are distinguished: appendage-dependent (swimming, swarming, and twitching) and appendage-independent (gliding and sliding). All of them allow bacteria to relocate and aid bacteria during infection. Swimming motility allows bacteria to spread from ‘persister cells’ in biofilm microcolonies and colonise other tissues. Twitching motility enables bacteria to press through the tissues during infection, whereas sliding motility allows cocci (defined as non-motile) to migrate over surfaces. Bacteria during swarming display greater resistance to antimicrobials. Molecular motors generating the focal adhesion complexes in the bacterial cell leaflet generate a ‘wave’, which pushes bacterial cells lacking appendages, thereby enabling movement. Here, we present the five main types of bacterial motility, their molecular mechanisms, and examples of bacteria that utilise them. Bacterial migration mechanisms can be considered not only as a virulence factor but also as a target for antibacterial therapy.
Skin and wound infections are serious medical problems, and the diversity of bacteria makes such infections difficult to treat. Bacteria possess many virulence factors, among which motility plays a key role in skin infections. This feature allows for movement over the skin surface and relocation into the wound. The aim is to review the type of bacterial movement and to indicate the underlying mechanisms than can serve as a target for developing or modifying antibacterial therapies applied in wound infection treatment. Five types of bacterial movement are distinguished: appendage-dependent (swimming, swarming, and twitching) and appendage-independent (gliding and sliding). All of them allow bacteria to relocate and aid bacteria during infection. Swimming motility allows bacteria to spread from ‘persister cells’ in biofilm microcolonies and colonise other tissues. Twitching motility enables bacteria to press through the tissues during infection, whereas sliding motility allows cocci (defined as non-motile) to migrate over surfaces. Bacteria during swarming display greater resistance to antimicrobials. Molecular motors generating the focal adhesion complexes in the bacterial cell leaflet generate a ‘wave’, which pushes bacterial cells lacking appendages, thereby enabling movement.
- swimming
- swarming
- twitching
- gliding
1. Introduction
2. Types of Bacterial Movement
One of the main characteristics of microbes adapting to and surviving in the external environment is how they move and grow, whether in liquid, semi-liquid, or solid environments. Notably, not all bacteria are capable of active movement. In most cases, the flagella enable motion. Bacterial cells have flagella in variable quantities and locations, which is one of the features that enable their classification. However, different movement phenomena, such as the result of type IV pili (T4P), are still being investigated. In 1972, the different types of microbial translocation were characterised and named [29][26]. Five of the original terms (swimming, swarming, twitching, gliding, and sliding) are still widely used [30][27]. Differences in bacterial types of movement are shown in Figure 1.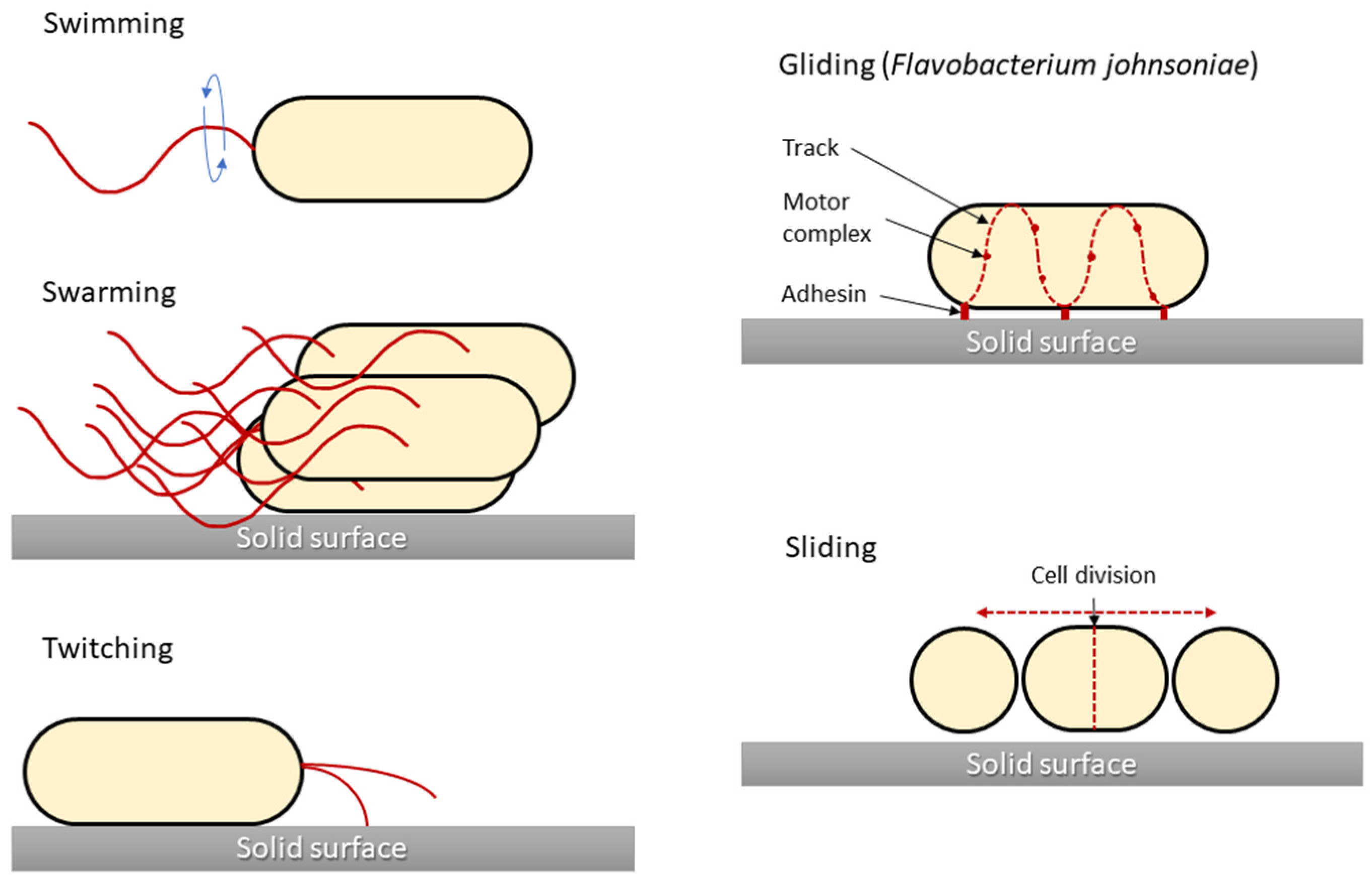
2.1. Swimming Motility
Swimming motility is one of the basic skills in the life cycle of bacteria, giving them an advantage in surviving unfavourable environmental conditions [32][28]. Swimming motility is an individual cell movement that uses flagella rotation to move through aqueous environments [33,34][29][30]. Flagellated bacteria typically swim in a series of more or less straight runs interrupted by short reorientations [35][31]. Each flagellum is rotated by a motor that is embedded in the cell membrane. Flagella are highly complex bacterial organelles that are unusually well conserved among diverse bacterial species. Over 60 structural and regulatory proteins are involved in flagellum synthesis and function [36][32]. There are three elements in the structure of flagella found in prokaryotes: the fibre, the hook, and the basal body. A cell may have one, two, or more flagella on the front or back of the cell, over part of it, or covering its entire surface. The outer part of the cilia consists of the flagellin protein, of which there are approximately 20,000 subunits in a single cilium. The flagellum is anchored in the bacterial cell by a basic body consisting of the L, P, S, M, and C rings surrounding the cylindrical part. Gram-positive bacteria do not have the two outer L and P rings, which distinguishes them from Gram-negative bacteria with five rings [37][33]. The flagella fibre is connected to the basic body with a flexible section called a hook, the length of which is approximately 55 nm [38][34]. The direction and regulation of flagellar rotation enable bacteria to move in chemical gradients, termed chemotaxis [35,39][31][35]. This complex behaviour begins in cell membrane chemoreceptors, which detect chemical compounds and respond to them by changing their conformation. E. coli has five such receptors (Tar, Tsr, Tap, Trg, and Aer), which are arranged in chemoreceptor clusters, together with two cytoplasmic proteins, the adaptor CheW and the histidine autokinase CheA [40][36]. Motility regulation by the chemotaxis system is well investigated for a variety of bacterial species. Chemical gradients are sensed by chemoreceptors, which trigger the autophosphorylation of the cytoplasmic CheA, which forms a complex with the receptor through the coupling protein CheW [33,41][29][37]. In response to these changes, CheA transfers its phosphate group to CheY, a diffusible cytoplasmic response regulator that interacts with the flagellar motor and leads to a modulation of motility characteristics [33,41][29][37]. This signalling core is highly conserved among all chemotaxis pathways [42][38]. Although only a fraction of bacteria associated with animal hosts are motile, flagellar motility and chemotaxis are important for the successful colonisation and virulence of many pathogens, for example, gastrointestinal Campylobacter jejuni, Salmonella enterica serovar Typhimurium, Helicobacter pylori, and Vibrio cholerae [41][37]. In several cases, it has also been shown that the same flagellin, the protein subunit comprising the bacterial flagellum, can be a major driver of inflammation [35][31]. Motility might have several functions in the host–microbe interactions. For example, in the early stages of biofilm formation, planktonic bacteria swim close to the surface by rotating their flagella and attaching to the surface using their pili [43][39]. The most common opportunistic pathogen in wound infections is P. aeruginosa. Many studies have demonstrated that P. aeruginosa biofilms are the key factors for exacerbating the skin inflammatory response and its resistance to antimicrobial agents [13,36][13][32]. Bacteria are inextricably associated with wound healing. However, the latest research indicates that they are not always associated with infection and worse wound healing. It has been reported that the skin’s natural microbiome can contribute to more rapid wound healing. Several studies were performed with mice infected with staphylococci. Interestingly, S. aureus was found to be the best healing inducer. Sometimes, swimming bacteria are used by non-flagellated bacteria that do not have the capacity to independently translocate with this mechanism. The genus Staphylococcus, for example, is classically considered non-motile in fluid environments due to a lack of flagella. Despite their motility limitations, staphylococcal species effectively reach and thrive in their preferred ecological niches. Data suggested that S. aureus has acquired, through P. aeruginosa, an increased capacity to travel longer distances, allowing it to colonise niches that are relatively inaccessible in the absence of swimming carrier bacteria, which may be due to the hitchhiking of S. aureus on P. aeruginosa. It was also observed that P. aeruginosa can carry another staphylococcal cargo, Staphylococcus epidermidis, which may also be important in wound infection [44][40].2.2. Swarming Motility
Swarming motility is mainly based on the differentiation of vegetative cells into swarming cells with a large number of flagella, which can migrate rapidly in a coordinated manner over solid surfaces. Bacteria display different phenotypes during swarming, but not all of them are equal: swarming P. aeruginosa do not develop typical swarmer cell phenotypes (elongated, hyper-flagellated cells) similar to other swarming bacteria, and individual cells swarming in rafts also reveal great variance and the lack of a highly differentiated swarmer cell phenotype [45][41]. Swarming phenotypes vary not only within a species but also within a strain, whereby the different P. aeruginosa strains grown on identical medium conditions will display different swarming patterns [46][42]. A swarm of migrating bacteria moves forward and traps a water reservoir, and in this moist region, individual cell speed is comparable to swimming speeds in bulk liquid, typically in the order of 20 μm/sec [47][43]. Bacterial swarming plays a crucial role in many pathogen–host interactions, and is considered an important virulence factor. The differentiation during swarming into the hyper-flagellated elongated cell of P. mirabilis is coupled to the ability of this bacterium to enter host cells, with the manifestation of upregulated virulence protein expression (haemolysin, urease, and protease) [48,49][44][45]. The stimuli that are necessary to control this movement are responsive to cell density, surface contact, and physiological signals. The swarming motility needs signals from the quorum sensing system and the cyclic di-GMP network that regulates the transitions between motile and biofilm modes across many bacterial species [50][46]. These signals activate flagella biogenesis via the main flagellar operon flhDC, which is the main point of the regulatory network of cell differentiation and migration and is crucial for swarming motility [48,51][44][47]. Surface sensing occurs in two proposed ways, through the inhibition of flagellar rotation and/or through the detection of the O antigen contact of lipopolysaccharide (LPS) with a solid surface [51][47] in Gram-negative bacteria. In addition, LPS is suggested to act as an osmolarity agent and facilitate swarming [52][48]. The swarming phenomenon is characterised by bacterial growth in the form of zones with clearly delimited darker peripheral circles on an agar medium. In a typical swarming, after contact with a surface, microbial cells are able to extend up to 40 times and increase the number of flagella. In the following stage, cells divide into short cells that are unable to move. The cycles of cell elongation, movement, and division are repeated constantly, resulting in a characteristic “pattern” of their growth on the substrate, in which the darker rings reflect the periods of bacterial division. Changes in cell length and shape result from changes in the hardness of the surface on which the cells grow, and the presence of LPS, which participates in the regulation of osmolarity, helps them overcome the environmental barriers that inhibit bacterial mobility. Among others, E. coli, Salmonella spp., P. mirabilis, Bacillus subtilis, and P. aeruginosa are able to migrate in this way. Compared with swimming, chemotaxis is unnecessary here, and the oscillating motion is mainly based on the flagellar drive and the mechanical interactions taking place [53][49]. The swarming motility is also closely associated with the resistance of bacterial cells, which are antibiotic-sensitive, providing cells with greater availability of nutrients and competitive advantage due to secreted surfactants [48][44]. For example, the multi-resistant phenotype of P. aeruginosa is closely associated with swarming growth and has a transient form [54][50]. Currently, research on this motility is focused on obtaining more detailed information about their lipids, proteins, and enzymes, as well as their relationships, the metabolic pathways involved, and methods of regulation at the molecular level [55][51].References
- Schommer, N.N.; Gallo, R.L. Structure and Function of the Human Skin Microbiome. Trends Microbiol. 2013, 21, 660–668.
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The Human Skin Microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155.
- DiNubile, M.J.; Lipsky, B.A. Complicated Infections of Skin and Skin Structures: When the Infection Is More than Skip Deep. J. Antimicrob. Chemother. 2004, 53, ii37–ii50.
- Kaye, K.S.; Petty, L.A.; Shorr, A.F.; Zilberberg, M.D. Current Epidemiology, Etiology, and Burden of Acute Skin Infections in the United States. Clin. Infect. Dis. 2019, 68, S193–S199.
- Tognetti, L.; Martinelli, C.; Berti, S.; Hercogova, J.; Lotti, T.; Leoncini, F.; Moretti, S. Bacterial Skin and Soft Tissue Infections: Review of the Epidemiology, Microbiology, Aetiopathogenesis and Treatment: A Collaboration between Dermatologists and Infectivologists. J. Eur. Acad. Dermatol. Venereol. 2012, 26, 931–941.
- Plewig, G.; French, L.; Ruzicka, T.; Kaufmann, R.; Hertl, M. Braun-Falco’s Dermatology, 2020th ed.; Springer: Berlin/Heidelberg, Germany, 2020.
- Cates, J.E.; Mitrani-Gold, F.S.; Li, G.; Mundy, L.M. Systematic Review and Meta-Analysis to Estimate Antibacterial Treatment Effect in Acute Bacterial Skin and Skin Structure Infection. Antimicrob. Agents Chemother. 2015, 59, 4510–4520.
- Olsson, M.; Järbrink, K.; Divakar, U.; Bajpai, R.; Upton, Z.; Schmidtchen, A.; Car, J. The Humanistic and Economic Burden of Chronic Wounds: A Systematic Review: The Humanistic and Economic Burden of Chronic Wounds. Wound Repair Regen. 2019, 27, 114–125.
- Bessa, L.J.; Fazii, P.; di Giulio, M.; Cellini, L. Bacterial Isolates from Infected Wounds and Their Antibiotic Susceptibility Pattern: Some Remarks about Wound Infection. Int. Wound J. 2015, 12, 47–52.
- Puca, V.; Marulli, R.Z.; Grande, R.; Vitale, I.; Niro, A.; Molinaro, G.; Prezioso, S.; Muraro, R.; di Giovanni, P. Microbial Species Isolated from Infected Wounds and Antimicrobial Resistance Analysis: Data Emerging from a Three-Years Retrospective Study. Antibiotics 2021, 10, 1162.
- Wolcott, R.D.; Hanson, J.D.; Rees, E.J.; Koenig, L.D.; Phillips, C.D.; Wolcott, R.A.; Cox, S.B.; White, J.S. Analysis of the Chronic Wound Microbiota of 2,963 Patients by 16S RDNA Pyrosequencing. Wound Repair Regen. 2016, 24, 163–174.
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539.
- Guoqi, W.; Zhirui, L.; Song, W.; Tongtong, L.; Lihai, Z.; Licheng, Z.; Peifu, T. Negative Pressure Wound Therapy Reduces the Motility of Pseudomonas Aeruginosa and Enhances Wound Healing in a Rabbit Ear Biofilm Infection Model. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2018, 111, 1557–1570.
- Mousavi, M.; Behrouz, B.; Irajian, G.; Mahdavi, M.; Korpi, F.; Motamedifar, M. Passive Immunization against Pseudomonas Aeruginosa Recombinant PilA in a Murine Burn Wound Model. Microb. Pathog. 2016, 101, 83–88.
- Pfalzgraff, A.; Brandenburg, K.; Weindl, G. Antimicrobial Peptides and Their Therapeutic Potential for Bacterial Skin Infections and Wounds. Front. Pharmacol. 2018, 9, 281.
- Nakagami, G.; Morohoshi, T.; Ikeda, T.; Ohta, Y.; Sagara, H.; Huang, L.; Nagase, T.; Sugama, J.; Sanada, H. Contribution of Quorum Sensing to the Virulence of Pseudomonas Aeruginosa in Pressure Ulcer Infection in Rats. Wound Repair Regen. 2011, 19, 214–222.
- Han, G.; Ceilley, R. Chronic Wound Healing: A Review of Current Management and Treatments. Adv. Ther. 2017, 34, 599–610.
- Huang, X.; Liang, P.; Jiang, B.; Zhang, P.; Yu, W.; Duan, M.; Guo, L.; Cui, X.; Huang, M.; Huang, X. Hyperbaric Oxygen Potentiates Diabetic Wound Healing by Promoting Fibroblast Cell Proliferation and Endothelial Cell Angiogenesis. Life Sci. 2020, 259, 118246.
- Agarwal, P.; Kukrele, R.; Sharma, D. Vacuum Assisted Closure (VAC)/Negative Pressure Wound Therapy (NPWT) for Difficult Wounds: A Review. J. Clin. Orthop. Trauma 2019, 10, 845–848.
- Gupta, S.; Gabriel, A.; Lantis, J.; Téot, L. Clinical Recommendations and Practical Guide for Negative Pressure Wound Therapy with Instillation. Int. Wound J. 2016, 13, 159–174.
- Kloth, L.C. Discussion: Advanced Technologies to Improve Wound Healing: Electrical Stimulation, Vibration Therapy, and Ultrasound-What Is the Evidence? Plast. Reconstr. Surg. 2016, 138, 105–106.
- Balakatounis, K.C.; Angoules, A.G. Low-Intensity Electrical Stimulation in Wound Healing: Review of the Efficacy of Externally Applied Currents Resembling the Current of Injury. Eplasty 2008, 8, e28.
- Ashrafi, M.; Alonso-Rasgado, T.; Baguneid, M.; Bayat, A. The Efficacy of Electrical Stimulation in Lower Extremity Cutaneous Wound Healing: A Systematic Review. Exp. Dermatol. 2017, 26, 171–178.
- Singh, M.R.; Saraf, S.; Vyas, A.; Jain, V.; Singh, D. Innovative Approaches in Wound Healing: Trajectory and Advances. Artif. Cells Nanomed. Biotechnol. 2013, 41, 202–212.
- Bekara, F.; Vitse, J.; Fluieraru, S.; Masson, R.; de Runz, A.; Georgescu, V.; Bressy, G.; Labbé, J.L.; Chaput, B.; Herlin, C. New Techniques for Wound Management: A Systematic Review of Their Role in the Management of Chronic Wounds. Arch. Plast. Surg. 2018, 45, 102–110.
- Henrichsen, J. Bacterial Surface Translocation: A Survey and a Classification. Bacteriol. Rev. 1972, 36, 478–503.
- Shrout, J.D. A Fantastic Voyage for Sliding Bacteria. Trends Microbiol. 2015, 23, 244–246.
- Minnullina, L.; Kostennikova, Z.; Evtugin, V.; Akosah, Y.; Sharipova, M.; Mardanova, A. Diversity in the Swimming Motility and Flagellar Regulon Structure of Uropathogenic Morganella Morganii Strains. Int. Microbiol. 2022, 25, 111–122.
- Corral, J.; Sebastià, P.; Coll, N.S.; Barbé, J.; Aranda, J.; Valls, M. Twitching and Swimming Motility Play a Role in Ralstonia Solanacearum Pathogenicity. Msphere 2020, 5, e00740-19.
- Miyata, M.; Robinson, R.C.; Uyeda, T.Q.P.; Fukumori, Y.; Fukushima, S.I.; Haruta, S.; Homma, M.; Inaba, K.; Ito, M.; Kaito, C.; et al. Tree of Motility—A Proposed History of Motility Systems in the Tree of Life. Genes Cells 2020, 25, 6–21.
- Colin, R.; Ni, B.; Laganenka, L.; Sourjik, V. Multiple Functions of Flagellar Motility and Chemotaxis in Bacterial Physiology. FEMS Microbiol. Rev. 2021, 45, fuab038.
- Li, Y.; Xia, H.; Bai, F.; Song, X.; Zhuang, L.; Xu, H.; Zhang, X.; Qiao, M. PA5001 Gene Involves in Swimming Motility and Biofilm Formation in Pseudomonas Aeruginosa. Microb. Pathog. 2020, 144, 103982.
- Terashima, H.; Kawamoto, A.; Morimoto, Y.V.; Imada, K.; Minamino, T. Structural Differences in the Bacterial Flagellar Motor among Bacterial Species. Biophys. Physicobiol. 2017, 14, 191–198.
- Bardy, S.L.; Ng, S.Y.M.; Jarrell, K.F. Prokaryotic Motility Structures. Microbiology 2003, 149, 295–304.
- Sun, E.; Liu, S.; Hancock, R.E.W. Surfing Motility: A Conserved yet Diverse Adaptation among Motile Bacteria. J. Bacteriol. 2018, 191, 5569–5576.
- Schauer, O.; Mostaghaci, B.; Colin, R.; Hürtgen, D.; Kraus, D.; Sitti, M.; Sourjik, V. Motility and Chemotaxis of Bacteria-Driven Microswimmers Fabricated Using Antigen 43-Mediated Biotin Display. Sci. Rep. 2018, 8, 9801.
- Schwanbeck, J.; Oehmig, I.; Groß, U.; Zautner, A.E.; Bohne, W. Clostridioides Difficile Single Cell Swimming Strategy: A Novel Motility Pattern Regulated by Viscoelastic Properties of the Environment. Front. Microbiol. 2021, 12, 715220.
- Wang, X.; Vu, A.; Lee, K.; Dahlquist, F.W. CheA-Receptor Interaction Sites in Bacterial Chemotaxis. J. Mol. Biol. 2012, 422, 282–290.
- Khong, N.Z.J.; Zeng, Y.; Lai, S.K.; Koh, C.G.; Liang, Z.X.; Chiam, K.H.; Li, H.Y. Dynamic Swimming Pattern of Pseudomonas Aeruginosa near a Vertical Wall during Initial Attachment Stages of Biofilm Formation. Sci. Rep. 2021, 11, 1952.
- Samad, T.; Billings, N.; Birjiniuk, A.; Crouzier, T.; Doyle, P.S.; Ribbeck, K. Swimming Bacteria Promote Dispersal of Non-Motile Staphylococcal Species. ISME J. 2017, 11, 1933–1937.
- Madukoma, C.S.; Liang, P.; Dimkovikj, A.; Chen, J.; Lee, S.W.; Chen, D.Z.; Shrout, J.D. Single Cells Exhibit Differing Behavioral Phases during Early Stages of Pseudomonas Aeruginosa Swarming. J. Bacteriol. 2019, 201, e00184-19.
- Morris, J.D.; Hewitt, J.L.; Wolfe, L.G.; Kamatkar, N.G.; Chapman, S.M.; Diener, J.M.; Courtney, A.J.; Leevy, W.M.; Shrout, J.D. Imaging and Analysis of Pseudomonas Aeruginosa Swarming and Rhamnolipid Production. Appl. Environ. Microbiol. 2011, 77, 8310–8317.
- Be’er, A.; Ariel, G. A Statistical Physics View of Swarming Bacteria. Mov. Ecol. 2019, 7, 9.
- Kearns, D.B. A Field Guide to Bacterial Swarming Motility. Nat. Rev. Microbiol. 2010, 8, 634–644.
- Walker, K.E.; Moghaddame-Jafari, S.; Lockatell, C.V.; Johnson, D.; Belas, R. ZapA, the IgA-Degrading Metalloprotease of Proteus Mirabilis, Is a Virulence Factor Expressed Specifically in Swarmer Cells. Mol. Microbiol. 1999, 32, 825–836.
- Yan, J.; Monaco, H.; Xavier, J.B. The Ultimate Guide to Bacterial Swarming: An Experimental Model to Study the Evolution of Cooperative Behavior. Annu. Rev. Microbiol. 2019, 73, 293–312.
- Morgenstein, R.M.; Szostek, B.; Rather, P.N. Regulation of Gene Expression during Swarmer Cell Differentiation in Proteus Mirabilis. FEMS Microbiol. Rev. 2010, 34, 753–763.
- Toguchi, A.; Siano, M.; Burkart, M.; Harshey, R.M. Genetics of Swarming Motility in Salmonella Enterica Serovar Typhimurium: Critical Role for Lipopolysaccharide. J. Bacteriol. 2000, 182, 6308–6321.
- Damton, N.C.; Turner, L.; Rojevsky, S.; Berg, H.C. Dynamics of Bacterial Swarming. Biophys. J. 2010, 98, 2082–2090.
- Lai, S.; Tremblay, J.; Déziel, E. Swarming Motility: A Multicellular Behaviour Conferring Antimicrobial Resistance. Environ. Microbiol. 2009, 11, 126–136.
- Poudel, S.; Giannone, R.J.; Farmer, A.T.; Campagna, S.R.; Bible, A.N.; Morrell-Falvey, J.L.; Elkins, J.G.; Hettich, R.L. Integrated Proteomics and Lipidomics Reveal That the Swarming Motility of Paenibacillus Polymyxa Is Characterized by Phospholipid Modification, Surfactant Deployment, and Flagellar Specialization Relative to Swimming Motility. Front. Microbiol. 2019, 10, 2594.
