Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Victoria V. Shumyantseva and Version 2 by Lindsay Dong.
The functional significance of cytochrome P450s (CYP) enzymes is their ability to catalyze the biotransformation of xenobiotics and endogenous compounds. P450 enzymes catalyze regio- and stereoselective oxidations of C-C and C-H bonds in the presence of oxygen as a cosubstrate. Initiation of cytochrome P450 catalytic cycle needs an electron donor (NADPH, NADH cofactor) in nature or alternative artificial electron donors such as electrodes, peroxides, photo reduction, and construction of enzymatic “galvanic couple”.
- cytochrome P450
- photocatalysis
- biocatalysis
- electron transfer
- electrochemistry
1. Introduction
Cytochrome P450s (CYPs) are unique enzymes with great transforming and synthetic activities. CYPs possess unique catalytic activities such as monooxygenase, oxygen reductase or substrate reductase. CYPs participate in chemical reactions of heteroatom oxygenation and dealkylation, aromatic and aliphatic hydroxylation, and cleavage of esters and oxidation of double bonds [1][2][3][1,2,3]. The P450 monooxygenase system is capable of metabolizing medicinal drugs, pollutants, carcinogens, and steroid hormones. Cytochrome P450 monooxygenases exhibit great potential for application in the role of bioreactors for the decomposition of a variety of hydrophobic chemicals, including pollutants. CYPs also possess stereo specificity for the biosynthetic application and synthesis of new drugs or metabolites with a new spectrum of pharmacological activities [4][5][4,5]. The development of new innovative approaches based on the functional role of CYPs is a prospective and promising method for the practical use of these enzymes in toxicology, pharmacology, new drug metabolism, and biomedical application. However, CYPs as hemoproteins cannot implement such a great variety of biotransformation reactions alone and need protein redox partners. Type I P450 catalytic system is a three-component one, in which the redox partners represent a FAD-containing ferredoxin reductase and iron-sulfur ([Fe-S]) protein ferredoxin. The mitochondrial and bacterial CYP systems belong to a Type I system. Type II P450 catalytic system is a two-component metabolic machine that consists of FAD/FMN-containing flavoprotein, NADPH-dependent cytochrome P450-reductase (CPR), and transfers the reducing equivalents to hemoprotein. Microsomal CYP systems are Type II (class II). Type III is a self-sufficient one-component system with a reductase and heme domain on one polypeptide chain [6][7][8][9][10][6,7,8,9,10].
2. Active Metals as Electron Donor for the Reduction in Heme Iron of CYPs
The application of active metals as alternative electron sources is a prospective approach in the construction of effective biocatalytic systems. Additional equipment is not necessary for active metal application. The system only needs the appropriate protein and active metal to supply the system with electrons. The starting step in the CYP electron transfer chain is the reaction of ferric (Fe3+) with one electron leading to the formation of ferrous (Fe2+) [4][9][10][4,9,10]. The operating principle of a galvanic element is based on the difference in the electrode potentials of the oxidant and the reducing agent. For (Fe3+) +1 electron → (Fe2+) reaction reduction potential is +0.77 V (vs. SHE, standard hydrogen electrode) [11][13]. Active metals (e.g., Zn, Mg, Ti) are the most commonly used as reducing agents, while hemoproteins are used as oxidants. Hemoproteins have a more positive standard electrode potential than zinc, such as E0 (Zn0/Zn2+) = −0.763 V; therefore, zinc serves as a reducing agent in a zinc-hemoprotein “biogalvanic element” (the NADPH/NADP+ cofactor redox potential is −0.32 V (vs. SHE) in aqueous media). Scheme 1 represents a relative range of metal activity based on redox potentials vs. SHE.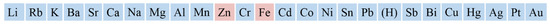
Scheme 1.
A relative range of metal activity vs. standard hydrogen electrode.
For modelling CYP-catalyzed reactions, an artificial hemoprotein was designed. Upon the complex formation of human serum albumin with iron protoporphyrine IX, there occurred the incorporation of heme into the protein and the formation of a specific complex with the albumin to heme molar ratio 2:1.
Cytochrome P450BM3 (CYP102A1) is Type III self-sufficient flavohemoprotein with heme and flavin domain on one polypeptide chain [2][3][12][2,3,18]. Cytochrome P450BM3 (CYP102) is a water-soluble, NADPH-dependent fatty acid hydroxylase from Bacillus megaterium that catalyzes the sub-terminal oxidation of saturated and unsaturated fatty acids. Several mutants of cytochrome P450BM3, such as P450BM3 M7 mutant (F87A V281G M354S R471C A1011T S1016G Q1022R) and P450BM3 M9 (R47F F87A M238K V281G M354S D363H W575C A595T) with improved electron transfer rate have been obtained [12][18]. P450BM3 M7 and P450BM3 M9 mutants were immobilized on DEAE-650S, further entrapped with k-carrageenan together with zinc dust, which functions as an electron source. For the effective reduction in CYP102 mutants, Zn/cobalt sepulchrate3+ complex (Zn/Co(III)sep) was used. Zn served as electron donor, Co(III)sep (S)-[1,3,6,8,10,13,16,19-octaazabicyclo-[6,6,6,]eicosane)cobalt(III)3+) served as electron transfer mediator.
3. Light-Driven CYP Catalysis
The aim of the elaboration of CYP-based systems with alternative electro sources is to avoid expensive NADPH cofactor and protein redox partners [2][3][7][2,3,7]. For the effective photochemical electron transfer, three main compounds should be present for the realization of the light-dependent P450 catalysis (Figure 1).
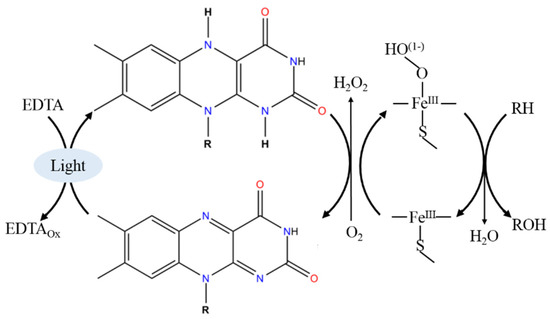

Figure 1.
Schematic representation of the light-driven heme iron reduction. R-substrate, ROH-metabolite of CYP-dependent reaction.
Light sources → Sacrificial electron donors → Photosensitizer → Heme iron
Different types of photosensitizers have been used for the light-driven reduction of CYP enzymes, such as biological Photosystem I, nanomaterials, quantum dots (CdS, TiO2, and Ag nanoparticles), prosthetic group analogs (deazaflavins, isoalloxazine-type flavins), organic dyes (eosin Y), inorganic metal complexes, such as ruthenium polypyridine ([Ru(bpy)3]2+), zinc porphyrin, and their derivatives [13][14][19,20].
The electron transfer cascade in the photosensitizers/protein complex may be realized as a covalent or non-covalent binding, permitting the variation of the mode of interaction [1][15][16][1,24,27].
Sacrificial electron donors (sacrificial reductants) include ascorbic acid, triethanolamine (TEOA), diethyldithiocarbamate (DTC), 2-(N-morpholino), ethanesulfonic acid (MES), and ethylenediaminetetraacetic acid (EDTA) [17][22].
For effective irradiation, light sources with different power and light emitting spectrum in the visible region are used as a source of energy, and varied distances from the reaction cell must also be the subject of discussion [12][13][14][15][17][18][19][20][18,19,20,21,22,23,24,25].
Reductase cofactors, such as flavin mononucleotide (FMN), flavin adenine dinucleotide (FAD), or riboflavin, can be reduced photochemically using ethylenediaminetetraacetic acid (EDTA) as an electron donor [12][18]. During photo activation, reduced flavins can generate hydrogen peroxide, so peroxide shunt pathway is realized [18][21].
Bacterial cytochromes P450BM3 (CYP CYP102), CYP199A4, from Rhodopseudomonas palustris HaA2 (as T252E mutant), algal P450s, artificial hemoprotein as a complex of human serum albumin and heme, reductase-free human CYPs expressed in Escherichia coli were used in photobiocatalytic systems [15][16][20][21][22][23][24][16,24,25,26,27,28,29].
Whole cells expressing in Escherichia coli human CYPs 1A1, 1A2, 1B1, 2E1, and 3A4 for the bioconversion of marketed drugs and steroids were conducted to demonstrate the general applicability of the photobiocatalytic system [23][25][17,28]. Substrate conversion was registered for 4-nitrophenol as model substrate and drugs chlorzoxazone, lovastatin, simvastatin, and 17β-estradiol. For whole-cell photocatalysis, the authors observed the transport of flavins into the Escherichia coli cells producing CYP2E1 by cytometric analysis based on fluorescent intensity assay.
4. Electrochemical Technology for Effective CYP Catalysis
Despite the great potential of cytochrome P450s, the dependence on expensive nicotinamide cofactor (NADPH) and protein redox partners, such as NADPH-P450 reductase (CPR), limits their employment in synthetic chemistry, pharmacology, nanobiotechnology and industry [1][2][3][7][8][26][1,2,3,7,8,11]. Since the catalytic cycle of cytochromes P450 is associated with the transfer of electrons [1][2][3][1,2,3], the use of electrochemical systems has found its practical application for modeling catalytic reactions of this class of hemoproteins. In electrochemical systems, electrodes can supply enzymes with electrons instead of NADPHs. For efficient electron transfer, the modification of electrodes and immobilization of enzymes on electrodes are necessary [27][28][29][30][30,31,32,33]. Electroanalytical methods demonstrate privileges such as high analytical sensitivity, the application of disposable or reusable electrodes that can be modified with a broad spectrum of nanocomposite materials to obtain smart electrodes, the development of both analytical and compact equipment with friendly software for the registration and analysis of the data obtained. Bioelectronics initiation of cytochrome P450s catalysis needs an appropriate type of electrode with a rational design of sensor modifiers [27][30]. Electrochemical methods are a modern, highly sensitive analytical tool for studying various functional aspects of cytochromes P450: the search for substrates, inhibitors, effectors, and activators as new potential drug candidates [28][29][30][31][32][33][34][35][31,32,33,34,35,36,37,38].4.1. Modelling on Electrode the Catalytic Cycle of CYP3A4
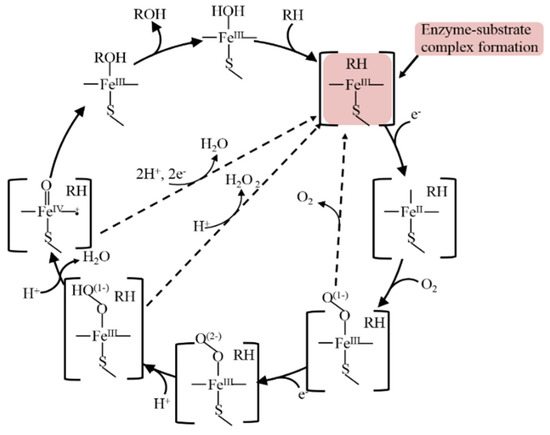
The biocatalytic mechanism of cytochrome P450s is very intricate and consists of several steps with intermediates formation [1][2][7][1,2,7]. The classical and well-known mechanism of CYP is represented in (Figure 23).
Figure 23.
A common catalytic cycle of CYPs enzymes.
Substrate (RH) binds to enzyme (E) with productive enzyme-substrate complex formation at the first step of CYP catalysis, with productive complex formation inducing a spin shift, which allows an Fe(III)-to-Fe(II) reduction [1][2][7][33][34][35][1,2,7,36,37,38]. The confirmation of the formation of an enzyme/substrate complex was first reaffirmed spectroscopically for the interaction of substrate 17-α-progesterone with an adrenal cortex microsomal enzyme [35][38].
4.2. Electron Transfer Chain Optimization on CYP-Electrode
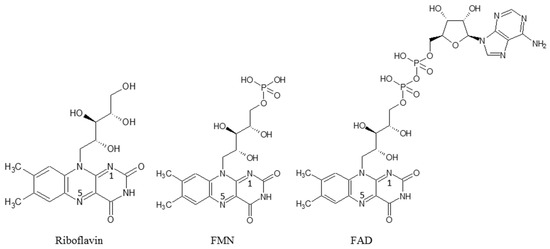
The electrochemical cytochrome P450 reactions have great potential for specific drug sensing, for searching for new drugs, and as bioreactors with broad synthetic applications. For the construction of a CYP-based electrochemical reactor, genetically engineered microsomes, human liver microsomes (HLM), and rat liver microsomes were used as the main participants of the electrochemical cell [36][37][50,51]. Microsomes consist of all proteins, which provide effective electron transfer to the heme iron ion for initiating the catalysis, such as cytochrome P450, and their redox partner protein CYP-NADPH reductase (CPR). Different types of electrodes were used for the microsomes’ utilization as bioreactors, such as a gold electrode, carbon electrode (polished basal plane pyrolytic graphite (BPG), edge plane pyrolytic graphite (EPG), glassy carbon (GC), or high-purity graphite (HPG) electrodes and modified with multiwalled carbon nanotubes EPG [36][37][50,51]. Rat liver microsomes (RLMs) were used for the detection of aflatoxin B1 (AFB1) metabolites, participating in carcinogenesis [38][52]. Electrochemical rat liver microsome-based biosensor using a composite of gold nanoparticles adsorbed on MXene (Au@MXene) for the rapid screening of AFB1. MXene is a new two-dimensional layered material-MXene which consists of transition metal carbides, nitrides, and carbonitrides. Rat liver microsomes (RLMs) were adsorbed on the Au@MXene nanocomposite and used for the detection of aflatoxin M1 in biosensor mode with a limit of detection of 2.8 nM. In spite of the effective substrate conversion, liver microsomes as bioreactors or biosensors possess ethical problems dealing with the liver as microsomes’ source. Fro this reason, artificial systems modeling the electron transfer chains of CYP-dependent microsomes were proposed [39][53]. Efficient work of mitochondrial and microsomal cytochrome P450 systems requires additional redox proteins (diflavin reductase and cytochrome b5). NADPH-dependent cytochrome P450 reductase contains both FAD and FMN as prosthetic molecules and belongs to the flavoproteins group [1][7][8][9][1,7,8,9]. The roles of flavin nucleotides are the coupling of the reaction of hydroxylation of substrates, an increase in the efficiency of enzyme catalysis, regulation of the flow of electrons, as well as stimulation of positive conformational changes in the structure of the protein [8][40][8,49]. The G. Gilardi group proposed the construction of effective electron transfer chains using the “Lego” approach, combining the heme domain of bacterial CYP102 A1 (BM3), CYP116B5 or CYP3A4 and the reductase domain of BM3 [41][42][43][44][45][46][47][48][49][58,59,60,61,62,63,64,65,66]. These constructs demonstrated enhanced efficiency in electrochemical systems. It was shown that interprotein electron transfer occurred from reduced flavin(s) to heme iron in flavohemeproteins [50][67]. Riboflavin was used as the model of reductase for the optimization and simplification of the electron transfer chain. In the presence of riboflavin as a mediator of electron flow and NADPH as an electron donor, bacterial types of cytochromes P450 CYP106A2, CYP107DY1, CYP107DY1, HmtS, HmtT, HmtN efficiently catalyzed the reaction of N-dealkylation of substrate diphenhydramine [51][68]. The efficiency of catalysis of covalent and non-covalent complexes of riboflavin as a simulator of flavoprotein and cytochrome P450 2B4 from rabbit liver in the presence of NADH was investigated earlier [52][53][69,70]. Based on these experiments, scholars used riboflavin, FMN and FAD as substitutes for reductase flavoprotein for the enhancement/improvement of electron transfer in electrochemical cytochrome P450 systems (Scheme 2).
Scheme 2.
Structures of flavin cofactors.
Hepatic enzyme cytochrome P450 3A4 (CYP3A4) is involved in the metabolism of about 50% of medicinal preparations and commercial drugs such as exogenous compounds. CYP3A4 catalyzes the metabolism of macrolide antibiotics (erythromycin, clarithromycin, azithromycin), calcium channel blockers (amlodipine, etc.), HIV protease inhibitors (indinavir, etc.), statins (such as simvastatin, atorvastatin), 5α-reductase inhibitors (finasteride), immunosuppressants (cyclosporine, etc.), antihistamines (astemizole), and prokinetics (cisapride) [5][6][5,6]. CYP3A4 also catalyzes the oxidation reactions of endogenous compounds, including estradiol (2- or 4-hydroxylation), testosterone (6β-hydroxylation), cortisol (6β-hydroxylation), cholesterol (4β-hydroxylation), progesterone (21-hydroxylation), cholic acid and chenodeoxycholic (formation of 3-dehydrocholic acid), and chenodeoxycholic (6α-hydroxylation) [5][6][5,6].
