Usually, in the early stages of the disease, the number of specific biomarkers is very low and sometimes difficult to detect using classical diagnostic methods. Among detection methods, biosensors have advantages such as easy operation, speed, and portability, with additional benefits of low costs and repeated reliable results. Single-molecule sensors such as nanopores that can detect biomolecules at low concentrations have the potential to become clinically relevant. As such, several applications have been introduced in this field for the detection of blood markers, nucleic acids, or proteins. The use of nanopores has yet to reach maturity for standardization as diagnostic techniques, however, they promise enormous potential, as progress is made into stabilizing nanopore structures, enhancing chemistries, and improving data collection and bioinformatic analysis.
- nanopore
- sensing
1. Introduction
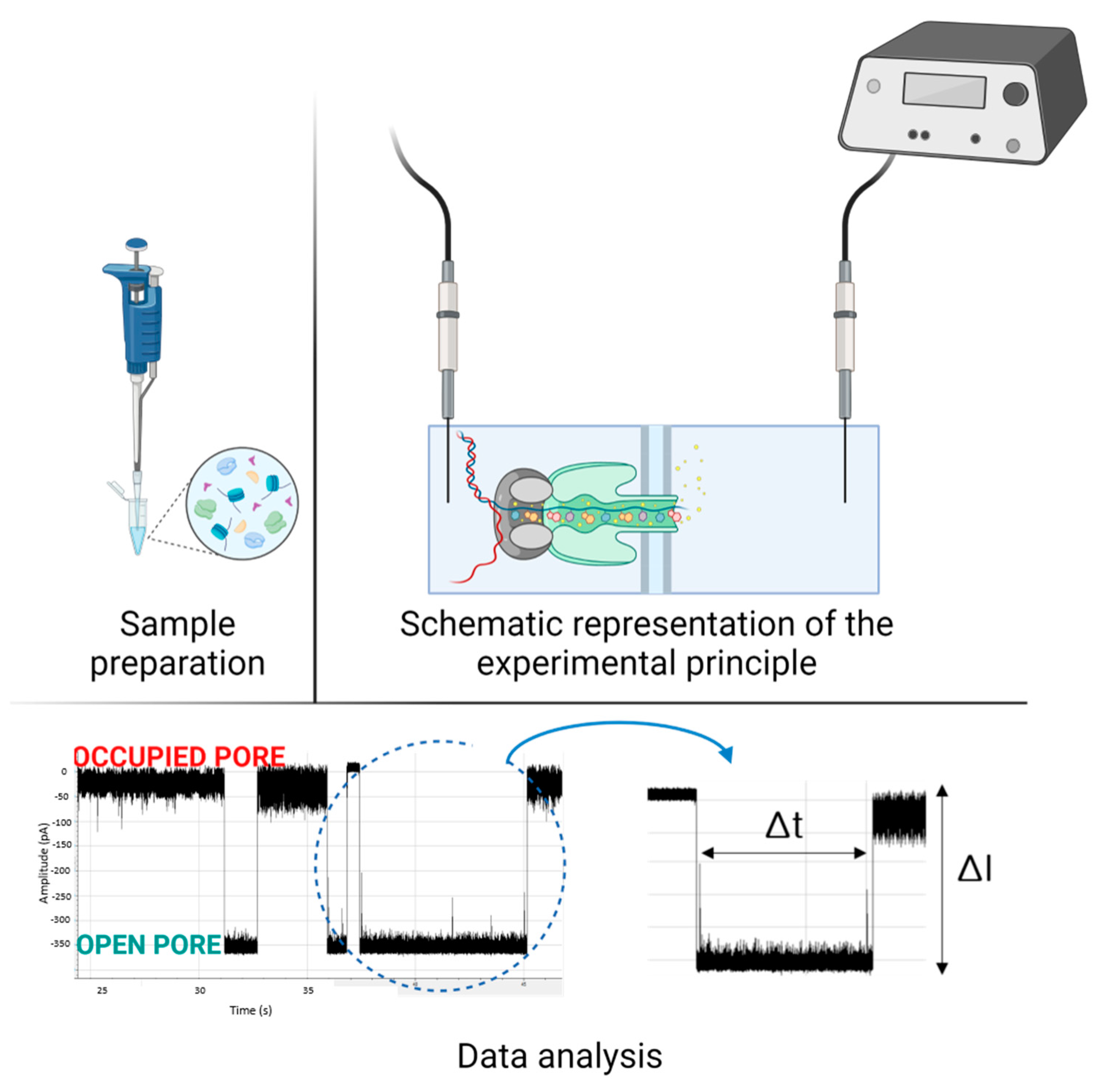
2. Applications in Sensing and Diagnostic
| Molecule of Interest |
Disease/Physiological Status | Experimental Setup | Reference | |
|---|---|---|---|---|
| Metabolic markers | Glucose | Diabetes, cancer | Glass nanopore based on nano pipettes | [22,23,24,25][19][20][21][22] |
| Vitamin B1 | Normal functioning of the human body | ClyA | [26][23] | |
| Uric acid | Gout, arthritis, kidney disease | Glass capillary nano-pipette | [27,28][24][25] | |
| Detection of nucleic acids | Bacteria | Infectious diseases | α-HL, silicon nanopore, MinION™ | [29,30,31,32][26][27][28][29] |
| Genetic markers: microRNA, pancreatic cancer marker, breast cancer marker | Cancer | Biological and solid-state nanopores | [33,34,35,36,37,38][30][31][32][33][34][35] | |
| Protein detection | Prion gene, ᏸ-amyloid, α-synuclein, PSA, insulin, hemoglobin | Neurologic diseases, cancer, autoimmune diseases, etc. | α-HL, ClyA, aerolysin, SiN, graphene Al2O3, SiO2, PlyAB | [17,39,40,41,42,43][14][36][37][38][39][40] |
References
- Kolmogorov, M.; Billingsley, K.J.; Meredith, M.; Monlong, J.; Lorig-Roach, R.; Asri, M.; Jerez, P.A.; Malik, L.; Dewan, R.; Reed, X.; et al. Scalable Nanopore sequencing of human genomes provides a comprehensive view of haplotype-resolved variation and methylation. bioRxiv 2023.
- Kono, N.; Arakawa, K. Nanopore sequencing: Review of potential applications in functional genomics. Dev. Growth Differ. 2019, 61, 316–326.
- Shi, W.; Friedman, A.K.; Baker, L.A. Nanopore Sensing. Anal. Chem. 2017, 89, 157–188.
- Smeets, R.M.M.; Keyser, U.F.; Krapf, D.; Wu, M.-Y.; Dekker, N.H.; Dekker, C. Salt Dependence of Ion Transport and DNA Translocation through Solid-State Nanopores. Nano Lett. 2005, 6, 89–95.
- Branton, D.; Deamer, D.W.; Marziali, A.; Bayley, H.; A Benner, S.; Butler, T.; Di Ventra, M.; Garaj, S.; Hibbs, A.; Huang, X.; et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008, 26, 1146–1153.
- Keyser, U.; Koeleman, B.N.; Van Dorp, S.; Krapf, D.; Smeets, R.M.M.; Lemay, S.G.; Dekker, N.; Dekker, C. Direct force measurements on DNA in a solid-state nanopore. Nat. Phys. 2006, 2, 473–477.
- Crnković, A.; Srnko, M.; Anderluh, G. Biological Nanopores: Engineering on Demand. Life 2021, 11, 27.
- Albrecht, T. Progress in single-biomolecule analysis with solid-state nanopores. Curr. Opin. Electrochem. 2017, 4, 159–165.
- Firnkes, M.; Pedone, D.; Knezevic, J.; Döblinger, M.; Rant, U. Electrically Facilitated Translocations of Proteins through Silicon Nitride Nanopores: Conjoint and Competitive Action of Diffusion, Electrophoresis, and Electroosmosis. Nano Lett. 2010, 10, 2162–2167.
- Nehra, A.; Ahlawat, S.; Singh, K.P. A biosensing expedition of nanopore: A review. Sens. Actuators B Chem. 2018, 284, 595–622.
- Li, J.; Golovchenko, J.A. Solid-State Nanopore for Detecting Individual Biopolymers. Micro Nano Technol. Bioanal. Methods Protoc. 2009, 544, 81–93.
- Gorzynski, J.E.; Goenka, S.D.; Shafin, K.; Jensen, T.D.; Fisk, D.G.; Grove, M.E.; Spiteri, E.; Pesout, T.; Monlong, J.; Baid, G.; et al. Ultrarapid Nanopore Genome Sequencing in a Critical Care Setting. N. Engl. J. Med. 2022, 386, 700–702.
- Goenka, S.D.; Gorzynski, J.E.; Shafin, K.; Fisk, D.G.; Pesout, T.; Jensen, T.D.; Monlong, J.; Chang, P.-C.; Baid, G.; Bernstein, J.A.; et al. Accelerated identification of disease-causing variants with ultra-rapid nanopore genome sequencing. Nat. Biotechnol. 2022, 40, 1035–1041.
- Luo, Y.; Wu, L.; Tu, J.; Lu, Z. Application of Solid-State Nanopore in Protein Detection. Int. J. Mol. Sci. 2020, 21, 2808.
- Ahmad, M.; Ha, J.-H.; Mayse, L.A.; Presti, M.F.; Wolfe, A.J.; Moody, K.J.; Loh, S.N.; Movileanu, L. A generalizable nanopore sensor for highly specific protein detection at single-molecule precision. Nat. Commun. 2023, 14, 1374.
- Wang, G.; Wang, L.; Han, Y.; Zhou, S.; Guan, X. Nanopore Stochastic Detection: Diversity, Sensitivity, and Beyond. Acc. Chem. Res. 2013, 46, 2867–2877.
- Bayley, H. Nanopore Sequencing: From Imagination to Reality. Clin. Chem. 2015, 61, 25–31.
- Yusko, E.C.; Bruhn, B.R.; Eggenberger, O.; Houghtaling, J.; Rollings, R.C.; Walsh, N.C.; Nandivada, S.; Pindrus, M.; Hall, A.R.; Sept, D.; et al. Real-time shape approximation and fingerprinting of single proteins using a nanopore. Nat. Nanotechnol. 2016, 12, 360–367.
- Mercado, R.; Moussy, F. In vitro and in vivo mineralization of Nafion membrane used for implantable glucose sensors. Biosens. Bioelectron. 1998, 13, 133–145.
- Li, Q.; Luo, G.; Feng, J.; Zhou, Q.; Zhang, L.; Zhu, Y. Amperometric Detection of Glucose with Glucose Oxidase Absorbed on Porous Nanocrystalline TiO2 Film. Electroanalysis 2001, 13, 413–416.
- Wei, A.; Sun, X.W.; Wang, J.X.; Lei, Y.; Cai, X.P.; Li, C.M.; Dong, Z.L.; Huang, W. Enzymatic glucose biosensor based on ZnO nanorod array grown by hydrothermal decomposition. Appl. Phys. Lett. 2006, 89, 123902.
- Saha, S.; Arya, S.K.; Singh, S.; Sreenivas, K.; Malhotra, B.; Gupta, V. Zinc oxide–potassium ferricyanide composite thin film matrix for biosensing applications. Anal. Chim. Acta 2009, 653, 212–216.
- Lucas, F.L.R.; Piso, T.R.C.; Heide, N.J.; Galenkamp, N.S.; Hermans, J.; Wloka, C.; Maglia, G. Automated Electrical Quantification of Vitamin B1 in a Bodily Fluid using an Engineered Nanopore Sensor. Angew. Chem. Int. Ed. 2021, 60, 22849–22855.
- He, H.; Xu, X.; Wang, P.; Chen, L.; Jin, Y. The facile surface chemical modification of a single glass nanopore and its use in the nonenzymatic detection of uric acid. Chem. Commun. 2014, 51, 1914–1917.
- Cao, S.; Ding, S.; Liu, Y.; Zhu, A.; Shi, G. Biomimetic Mineralization of Gold Nanoclusters as Multifunctional Thin Films for Glass Nanopore Modification, Characterization, and Sensing. Anal. Chem. 2017, 89, 7886–7892.
- Tang, Y.; Li, Z.; Luo, Q.; Liu, J.; Wu, J. Bacteria detection based on its blockage effect on silicon nanopore array. Biosens. Bioelectron. 2016, 79, 715–720.
- Cheng, M.S.; Lau, S.H.; Chow, V.T.; Toh, C.-S. Membrane-Based Electrochemical Nanobiosensor for Escherichia coli Detection and Analysis of Cells Viability. Environ. Sci. Technol. 2011, 45, 6453–6459.
- Tourancheau, A.; Mead, E.A.; Zhang, X.-S.; Fang, G. Discovering multiple types of DNA methylation from bacteria and microbiome using nanopore sequencing. Nat. Methods 2021, 18, 491–498.
- Schmidt, K.; Mwaigwisya, S.; Crossman, L.C.; Doumith, M.; Munroe, D.; Pires, C.; Khan, A.M.; Woodford, N.; Saunders, N.J.; Wain, J.; et al. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J. Antimicrob. Chemother. 2016, 72, 104–114.
- Wang, Y.; Zheng, D.; Tan, Q.; Wang, M.X.; Gu, L.-Q. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat. Nanotechnol. 2011, 6, 668–674.
- Xi, D.; Li, Z.; Liu, L.; Ai, S.; Zhang, S. Ultrasensitive Detection of Cancer Cells Combining Enzymatic Signal Amplification with an Aerolysin Nanopore. Anal. Chem. 2017, 90, 1029–1034.
- Liu, L.; Li, T.; Zhang, S.; Song, P.; Guo, B.; Zhao, Y.; Wu, H.-C. Simultaneous Quantification of Multiple Cancer Biomarkers in Blood Samples through DNA-Assisted Nanopore Sensing. Angew. Chem. Int. Ed. 2018, 57, 11882–11887.
- Bhatti, H.; Lu, Z.; Liu, Q. Nanopore Detection of Cancer Biomarkers: A Challenge to Science. Technol. Cancer Res. Treat. 2022, 21, 153303382210766.
- Henry, N.L.; Hayes, D.F. Cancer biomarkers. Mol. Oncol. 2012, 6, 140–146.
- Alzghoul, S.; Hailat, M.; Zivanovic, S.; Que, L.; Shah, G.V. Measurement of serum prostate cancer markers using a nanopore thin film based optofluidic chip. Biosens. Bioelectron. 2016, 77, 491–498.
- Stefureac, R.; Long, Y.T.; Kraatz, H.B.; Howard, P.; Lee, J.S. Transport of α-Helical Peptides through α-Hemolysin and Aerolysin Pores. Biochemistry 2006, 45, 9172–9179.
- Soskine, M.; Biesemans, A.; Moeyaert, B.; Cheley, S.; Bayley, H.; Maglia, G. An Engineered ClyA Nanopore Detects Folded Target Proteins by Selective External Association and Pore Entry. Nano Lett. 2012, 12, 4895–4900.
- Mereuta, L.; Roy, M.; Asandei, A.; Lee, J.K.; Park, Y.; Andricioaei, I.; Luchian, T. Slowing down single-molecule trafficking through a protein nanopore reveals intermediates for peptide translocation. Sci. Rep. 2014, 4, 3885.
- Freedman, K.J.; Bastian, A.R.; Chaiken, I.; Kim, M.J. Solid-State Nanopore Detection of Protein Complexes: Applications in Healthcare and Protein Kinetics. Small 2012, 9, 750–759.
- Turko, I.V.; Murad, F. Protein Nitration in Cardiovascular Diseases. 2002. Available online: http://pharmrev.aspetjournals.org (accessed on 3 June 2022).
