Regulation of the human IGF2 gene displays multiple layers of control, which secures a genetically and epigenetically predetermined gene expression pattern throughout embryonal growth and postnatal life. These predominantly nuclear regulatory mechanisms converge on the function of the IGF2-H19 gene cluster on Chromosome 11 and ultimately affect IGF2 gene expression. Deregulation of such control checkpoints leads to the enhancement of IGF2 gene transcription and/or transcript stabilization, ultimately leading to IGF-II peptide overproduction. This type of anomaly is responsible for the effects observed in terms of both abnormal fetal growth and increased cell proliferation, typically observed in pediatric overgrowth syndromes and cancer.
- IGF2, insulin-like growth factor 2 gene
- mRNA transcript
- IGF-II, insulin-like growth factor-2 peptide
1. IGF2 Gene Regulation at the Promoter and Transcript Level: An Unexploited View
2. The Human IGF2 Gene Structure
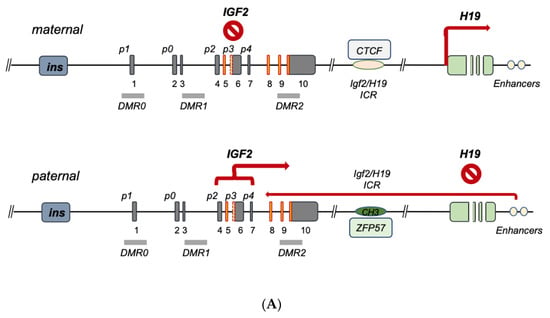
The human IGF2 gene occupies the 11p15.5 chromosomal locus, positioned between the insulin gene and the H19 gene, with which it establishes an imprinted gene cluster (NCBI Ref Seq NG_008849.1) [8][9] (Figure 1). The IGF2 gene is composed of 10 exons [9][10][11][10,11,12] whose expression is driven by five promoters (p0–p4), differentially activated from embryonal to postnatal life (see Table 2) [12][13][14][13,14,15]. The IGF2 gene product is a peptidic ligand (IGF-II), which plays a central role in embryonal growth in mammalians [15][16]. Furthermore, the role of secreted IGF-II autocrine and paracrine effects in tumorigenesis [15][16] and its growing role towards malignant feature maintenance are well documented (reviewed by Scalia et al. [16][17]). Interestingly, an alternatively expressed exonic region has been recently described as part of exon 6, and it appears to bear a role in diabetes predisposition (see Figure 1) [17][18]. For this reason, the understanding of IGF2 gene expression and transcriptional regulation bears intrinsic high biological and biomedical value. The IGF2 gene has been widely studied for its epigenetic parental (allele)-specific control. The established evidence demonstrates that in the majority of adult tissues, IGF2 is exclusively expressed by the paternal (methylated) allele due to its imprinting on the maternal (hypomethylated) allele, which is silenced as a result of its hypo-methylation status. In particular, the IGF2 promoter-specific differentially methylated regions (DMRs 0, 1, 2) partially overlap the IGF2 intronic and exonic sequences, along with the DMR known as “inter-genic- or IG-DMR” or Imprinting Center Region 1, ICR1. This region is located between the IGF2 and H19 genes coding regions and the IGF2 enhancer region downstream from H19, cumulatively establishing a phylogenetically conserved gene cluster acting as an epigenetic switch [18][19][20][19,20,21]. IG-DMR is an allele-dependent DMR (ICR1) containing the binding motif for the epigenetic master regulator CTCF, which, along with the PRC2 complex components (disucussed in Section 2 and summarized in Table 1). The CTCF–PRC2 complex binds the maternal hypomethylated ICR and insulates the IGF2 promoters [21][22][22,23]. On the contrary, the paternal ICR1, being prevented from CTCF binding as a consequence of ICR1 methylation status, results in a fully receptive effect of the enhancer regions, thereby displaying the classic monoallelic expression of the human imprinted IGF2 locus. This is graphically summarized in Figure 2A. A parallel promoter activation pattern for IGF2 expression in fetal growth, compared to the postnatal and adult phases, includes the promoter usage switching from the imprinted “fetal” (p2–p4) and “placental” (p0) promoters to the adult (p1) promoter [8][9]. Indeed, both cumulative and recent findings display a more diversified landscape of IGF2 regulation, extending beyond the previously known abnormalities linked to either (a) epigenetic deregulation or (b) allelic (uniparental) disomy, both of which are described in IGF2 overgrowth syndromes [23][24].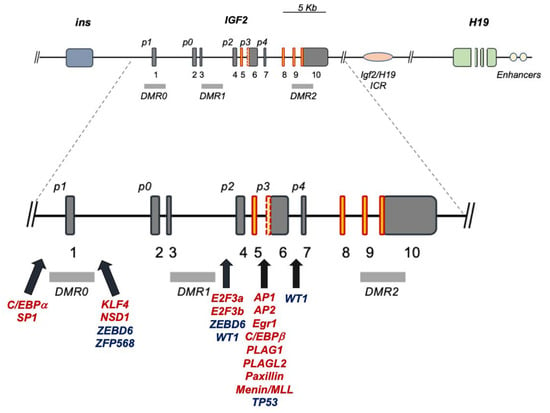
3. IGF2 Gene Regulation during Development and IGF2 Overexpression Syndromes
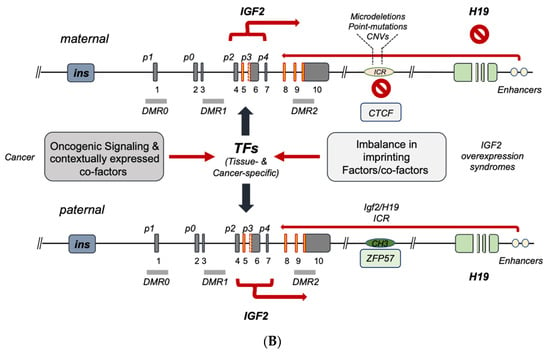
Seminal studies have shown the importance of genomic imprinting for the IGF2 gene and the entire IGF2-H19 gene cluster (reviewed in [8][9]). As discussed in the previous section, ICR1 differentially methylated status affects the binding of epigenetic master-regulator CTCF to unmethylated ICR1 motifs acting as an insulator [2][21][24][25][26][27][2,22,25,26,27,28]. More recently, the role of the imprinting factor ZFP57 on the methylation maintenance status of the paternal allele has been shown [28][29]. The above control model for IGF2 imprinting on the maternal and paternal alleles is schematically summarized in Figure 2A. The term ‘IGF2 overexpression syndromes’ relates to a variety of genetic abnormalities sharing the phenotype reported by Beckitt–Wiedemann to describe the resulting pediatric syndrome. A majority of the overgrowth symptoms in these subjects are secondary to the high levels of IGF-II produced at the embryonal and postnatal levels. This overproduction is mostly linked to the biallelic expression of IGF2 as a result of the imprinting relaxation of the maternal allele. A recent analysis of the genetic abnormalities in these subjects [23][24], leading to increased IGF2 activation, allows one to functionally classify the currently known IGF2 gene expression defects into two types, namely, (a) ICR defects causing the above relaxation on the maternal allele (via microdeletions and/or to DMR point mutations) [22][29][30][31][23,30,31,32], and (b) quantitative defects affecting the overall paternal gene cluster as a result of either uniparental disomy (UPD) or copy number variations (CNVs) [29][32][33][30,33,34]. An additional layer of control on IGF2 imprinting disclosed by recent studies relates to allele/DMR-specific factors acting as intrinsic-enabling factors and/or acting in synergy with CTCF on the maternal ICR [34][35]. Among these are Sox2/Oct4 [35][36][36,37], SUZ12 [37][38] and Vigilin [38][39], whose contextual functions as imprinting factors are governed by histone post-translational modifications, as evidenced by studies confirming their role in affecting both general and specific IGF2 imprinting. In particular, histone acetylation has been recognized since the late 1990s as a regulator of IGF2 imprinting, as shown by the ability of histone deacetylase inhibitors to cause IGF2 biallelic expression [39][40]. Even more detailed is the demonstration of the key role of H3K27 histone methylation for the proper maintenance of the maternal imprinting status via its effects on (a) the IGF2-H19 cluster loop conformation and (b) the DNA protein complex formation on the imprinted maternal allele [40][41]. In fact, in those cells with loss of imprinting (LOI), H3K27 demethylation leads to loss of the CTCF-orchestrated intrachromosomal loop between the IGF2 promoters and the ICR. The H3K27 methylation-free IGF2 promoters appear to become activated similarly to the paternal promoters, leading to biallelic expression. Noteworthily, SUZ12 has been shown to play a key role in the maintenance of the hypermethylation status of H3K27 by EZH2 since, in the absence of SUZ12, the PRC2 cannot be recruited to the maternal IGF2 promoter where this methylation takes place in order to induce the imprinting loop conformation [40][41]. The chromatin conformation at the IGF2-H19 cluster locus has been found to be essential for proper IGF2 expression, and this higher-order chromatin organization function is mediated by Cohesin [41][42]. Altogether, these studies point at a wider molecular network for the allele-specific control of IGF2 imprinting and offer additional potential mechanisms of dysregulation that could be responsible for those, yet unaccounted, molecular defects, leading to IGF2 increased transcription underlying the pathologic conditions discussed herein. A graphic summary of the human IGF2-H19 cluster regulation focusing on the latest landscape provided by the reviewed literature is conveyed in Figure 2 and Table 1.| Imprinting Factor | Key Feature | Reference(s) |
|---|---|---|
| CTCF | binds maternal ICR and insulates IGF2-p activity |
[34][35] |
| Cohesin | Cohesin is required for chromatin function at the H19/IGF2 locus | [41][42] |
| EZH2 | CH3-transferase component of PRC2 | [40][41] |
| SUZ12 | PRC2 component enabling ICR imprinting |
[37][38] |
| Sox2/Oct3–4 | CTCF-like effect | [35][36] |
| Vigilin | ICR imprinting effect via CTCF binding | [38][39] |
| ZFP57 | Binds paternal ICR and maintains methylated status | [28][29] |
4. IGF2 Gene Transcriptional Control in Cancer
A number of studies focusing on the role of IGF2 gene methylation and promoter usage in cancer have established the importance of IGF2 LOI status [42][43][44][45][43,44,45,46]. Nonetheless, the mechanistic relationship between promoter usage, both under monoallelic (under maintenance of imprinting, MOI) and biallelic status (caused by LOI), and the observed total IGF2 expression pattern/levels in cancer remains an active area of investigation. Indeed, a number of studies have shown a predominant activation of IGF2 fetal promoters (p2–p4) in a variety of cancers displaying IGF2 increased expression levels, with variable uncoupling of DMR0–2 methylation, along with monoallelic IGF2 and/or H19 expression [46][47][48][49][50][51][52][53][54][55][47,48,49,50,51,52,53,54,55,56]. In this context, it is important to stress that promoter usage and transcriptional activity are directly dependent on the involved transcription machinery, which is affected, in its turn, by the contextual transcriptional co-activator and co-repressor effects (also provided by the underlying IGF2 epigenetic protein–DNA interactions). In addition to the protein/DNA-driven control layer (or lack of control) of IGF2 gene expression, it is important to add the regulation layer provided by the RNA transcript control. This type of integrated approach to study IGF2 gene regulation both in IGF2 expression syndromes and in cancer, according to the authors of the present work, is essential in order to move the field beyond the historical (and still ongoing) compartmentalized approach to IGF2 gene studies. A graphic summary of our currenthe understanding of the regulation of the human IGF2 gene, spanning from IGF2 expression syndromes to cancer (overlapping in vivo), is provided in Figure 2B and Table 2.| Promoter Usage | Imprinting Control | Reference(s) |
|---|---|---|
| IGF2-p0 | Not imprinted Mostly active in fetal placenta |
[14][15] |
| IGF2-p1 | Not imprinted—mostly active in postnatal Liver |
[56][57][57,58] |
| IGF2-p2 | Imprinted- Mostly active during Fetal growth |
[58][59] |
| IGF2-p3 & IGF2-p3/p4 (*) |
Imprinted- Mostly active during Fetal growth, Widely reactivated in cancer |
[50][56][57][59][51,57,58,60] |
