Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Fanny Huang and Version 1 by Wangyang Meng.
Aberrant translation, a characteristic feature of cancer, is regulated by the complex and sophisticated RNA binding proteins (RBPs) in the canonical translation machinery. N6-methyladenosine (m6A) modifications are the most abundant internal modifications in mRNAs mediated by methyltransferase-like 3 (METTL3). METTL3 is commonly aberrantly expressed in different tumors and affects the mRNA translation of many oncogenes or dysregulated tumor suppressor genes in a variety of ways.
- METTL3
- m6A
- eukaryotic initiation factor
- cancer
1. Introduction
N6-methyladenosine (m6A) modifications in RNA were first identified in the 1870s [1]. The enzyme that catalyzes the formation of m6A is known as m6A “writer”, methyltransferase-like 3 (METTL3), which is the only catalytic subunit of the methyltransferase complex and can synthesize almost all m6A modifications in mRNAs [2]. As research progressed, it was gradually realized that m6A modifications are an essential regulatory modality in biological development, affecting cell differentiation and other physiological processes. Since the rise of m6A high-throughput sequencing methods in 2012 [3[3][4],4], more and more studies have found that the epitranscriptome plays a key role in regulating the fate and function of mRNAs in cells.
m6A is a selective modification enriched in specific mRNAs [5]. Some mRNAs contain only a single m6A site, but some contain 20 or even more m6A sites [4]. Overall, about 50–80% of mammalian mRNAs may m6A sites be absent [4,6,7,8][4][6][7][8]. Under physiological conditions, m6A is enriched in the 3’ untranslated region (3’UTR) and near the stop codons of the transcripts [3]. Analysis of mRNAs enriched in m6A modifications showed enrichment of developmental regulation and cell fate-related genes [9]. In contrast, transcripts of some highly stable “housekeeping” genes, including ribosomal proteins, showed a de-enrichment of m6A [9]. However, in pathological situations, some m6A sites may be regulated in a disease-specific manner. In various cellular stresses, the investigators also observed changes in m6A levels in the 5’UTR as well [10]. Thus, the role of m6A modifications in different diseases and the role they play remains to be elucidated.
In recent years, the role of m6A modifications in cancer has received increasing attention as epigenomics and oncology studies continue to progress. It has been found that m6A modifications in tumors can regulate the stability, splicing, nuclear translocation, and translation efficiency of various mRNAs [11[11][12],12], which in turn leads to a complex series of molecular events. m6A is added to RNA by the m6A writer-complex which includes METTL3, METTL14, WTAP, VIRMA, RBM15/15B, ZC3H13, and CBLL1 [13,14][13][14]. m6A readers include YTHDF1/2/3, YTHDC1/2, IGF2BP1/2/3, HNRNPC/G/A2B1 [13]. They act as RNA binding proteins to exert their effect on the RNA life cycle subsequently. m6A erasers, ALKBH5 and FTO, are demethylases that can remove m6A from RNA. As the key catalytic subunit forming m6A modification, there is increasing evidence in recent years that the m6A writer METTL3 is significantly aberrantly expressed in tumors and can play a key role as an oncogene in most cases, leading to different phenotypic changes in tumors, resulting in proliferation, invasion, metastasis, and drug resistance. For example, in bladder cancer, METTL3 is significantly overexpressed and is associated with proliferation, invasion, and tumorigenic capacity in in vivo, and METTL3 promotes tumor progression through m6A modification on AFF4 and NF-κB mRNA, which in turn activates MYC transcription [15]. In hepatoblastoma, abnormally high expression of METTL3 leads to a significant increase in m6A levels in the tumor, and m6A is enriched not only near the mRNA stop codon but also in the coding sequence (CDS) region. The elevated stability of CTNNB1 due to its m6A modification leads to significant activation of the Wnt/β-catenin signaling pathway, which in turn promotes malignant proliferation of tumors [16]. In esophageal cancer, METTL3 is also significantly overexpressed and can lead to mRNA degradation by upregulating the m6A level of APC mRNA and recruiting the m6A “reader” protein YTHDF2. The reduced expression of APC leads to abnormal activation of the Wnt/β-catenin signaling pathway, thus promoting the glycolytic process and malignant cell proliferation in tumors [17]. Interestingly, sometimes METTL3 also acts as a tumor suppressor. Cui et al. [18] reported that knocking down METTL3 altered m6A enrichment on ADAM19 and promoted the malignancy of glioblastoma stem cells. Wu et al. revealed that METTL3 mediated m6A modification of FBXW7 and suppressed the development of lung adenocarcinoma subsequently [19]. Thus, METTL3 regulates the fate of these RNAs through m6A modification at key transcripts, which in turn affects the development of many cancers, including hematologic malignancies and solid tumors.
2. Aberrant Translation in Cancer
In the life cycle of mRNA, translation, the process of protein synthesis, is the most energy-consuming step in the entire cell [20], and this step plays a key role in the regulation of gene expression. With the rapid development of high-throughput sequencing technologies in recent years, mathematical modeling and multi-omics analysis have revealed that the magnitude of translational regulation in cells exceeds the sum of transcription, mRNA degradation, and protein degradation [21]. Components of the translational machinery integrate almost all oncogenic signals [22], and dysregulation of the translational process is considered one of the hallmarks of tumors and is associated with abnormal proliferation, angiogenesis, differentiation, and immune response [23,24][23][24]. Aberrant mRNA translation is a common feature of tumors, in which the process cannot be separated from the involvement of RNA-binding proteins (RBPs) in canonical translation machinery, including eukaryotic initiation factor (eIF) and elongation factor (eEF) (Figure 1). Their signals are aberrantly amplified in tumors [25,26][25][26].
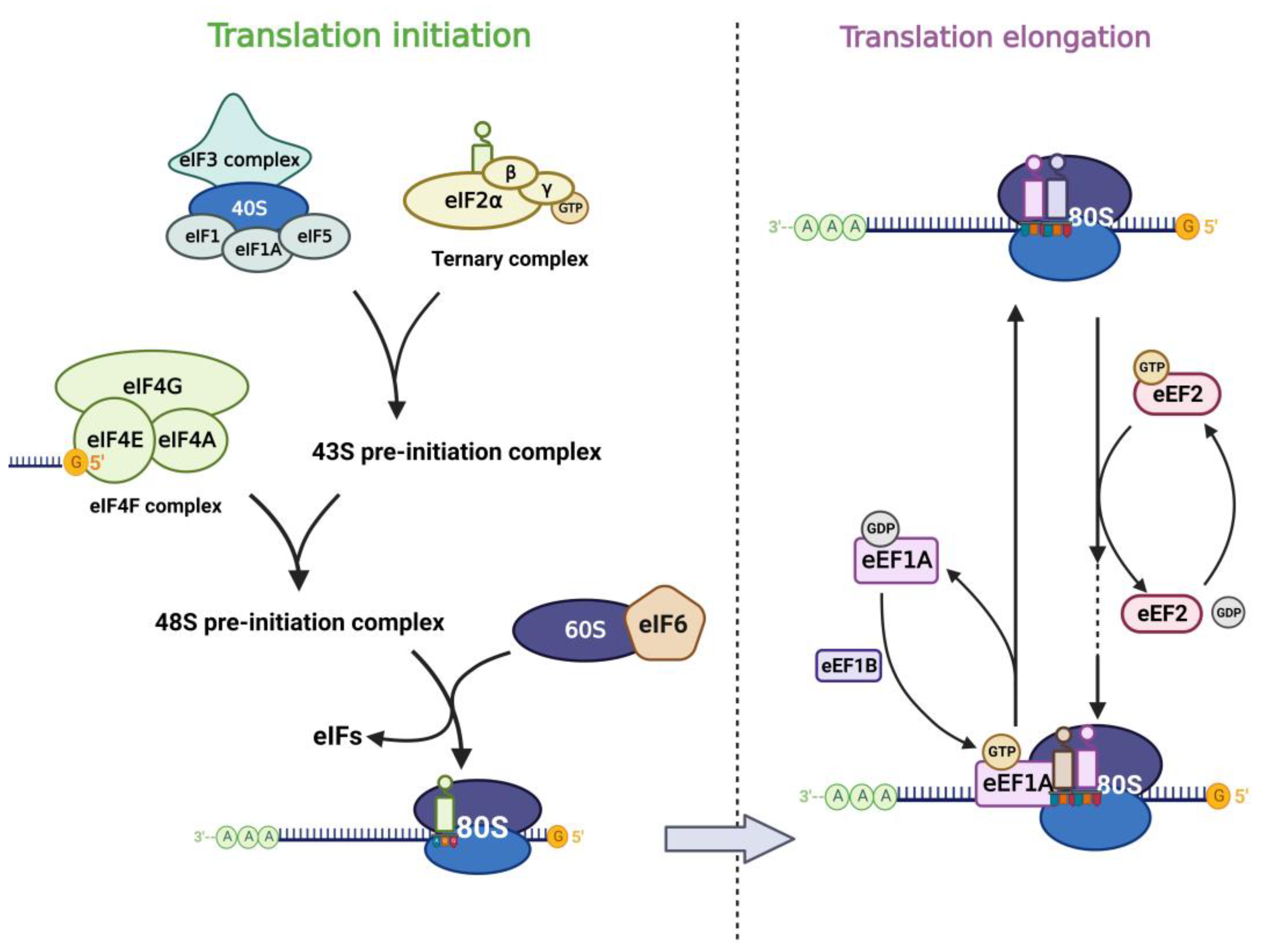
Figure 1. Translation factors participate in the mRNA translation process. The left panel shows the translation initiation process: eIF2 subunits combine with initiator methionyl tRNA and GTP to form the ternary complex (TC). TC associates with the 40S ribosomal subunit complex which consists of eIF3, eIF1, eIF1A, and eIF5 to form the 43S pre-initiation complex (43S PIC). Then 43S PIC is recruited to the mRNA template by combining to eIF4F complex and they form the 48S pre-initiation complex (48S PIC). The phosphorylation of eIF6 allows the 60S ribosomal subunit to join the 40S subunit, which leads to the formation of the translation-competent 80S ribosome and marks the end of translation initiation. The right panel shows the translation elongation cycle: eEF1A-GTP helps to deliver the aa-tRNA to the Aminoacyl site in ribosome and eEF1B recycles the released eEF1A-GDP subsequently. eEF2-GTP mediates the translocation of the elongating peptide to the Peptidyl site of the ribosome.
2.1. Translation Factors in Eukaryotic Translation
Translation of mRNA in eukaryotic cells includes cap-dependent and cap-independent translation, in which the eIF4F complex plays an important role. eIF4F contains three components (eIF4E, eIF4G, eIF4A), of which eIF4E is the cap-binding subunit of the eIF4F complex and is required for cap-dependent translation of all nuclear-encoded mRNAs [27]. In addition, eIF4E can also stimulate the RNA unwinding enzyme activity of eIF4A independently of its cap-binding function and thus promotes translation [28]. eIF4E interacts with eIF4G and binds the m7G cap structure of mRNA, which in turn promotes translation. eIF3 plays a central role in the translation initiation of classical cap-dependent translation and cap-independent translation [29,30,31][29][30][31]. Different subunits of eIF3 confer different functions to the eIF3 core complex. Besides, other eukaryotic initiation factors also play important role in the translation process. Translation initiation is generally regulated by the 43S pre-initiation complex (43S PIC), which consists of eIF1, eIF1A, eIF3, eIF5, and the ternary complex (TC) [22]. The TC is formed by eIF2 (containing α, β, γ subunits), tRNA, and GTP. When eIF2α is phosphorylated under stress, the TC formation is inhibited and the global translation is downregulated subsequently [32,33][32][33]. eIF6 was first reported to participate in the biogenesis of the 60S ribosomal subunit in the nucleus as an anti-association factor [34,35][34][35]. However, Gandin et al. [36] found that eIF6 is rate-limiting for efficient translation initiation. In the cytoplasm of mammalian cells, the phosphorylation of eIF6 on Ser235 leads to its release from the 60S, which promotes the formation of a translation-competent 80S ribosome [37]. The translation elongation process is carried out by the ribosome with the assistance of eEFs. Among them, eEF1A is an important component of the translational apparatus as it interacts with tRNA [26]. eEF2 possesses an RNA binding site that interacts with tRNAs and promotes conformational changes, thus allowing the latter to interact with the coding region of mRNAs, mediating the translocation of peptide chains in extension to the P-site of the ribosome [38].
2.2. Dysregulation of Translation Factors in Cancer
Most of the eIF4E-sensitive mRNAs have a long and highly structured 5’UTR region [39], and these mRNAs encode many proteins associated with cell proliferation and tumor progression, including MYC, VEGF, cyclin, and others [40]. Many studies have reported that overexpression of eIF4E is associated with poor prognosis in cancer patients and can lead to tumor vascularization and invasion [41]. In recent years, various subunits of eIF3 have been found to have altered expression in malignant tumors, affecting translation of oncogenic mRNAs. eIF3a expression level were first found to be elevated in breast cancer tissues compared to paired normal breast tissues by Bachmann et al. [42], and eIF3a might play an important role in regulating translation of specific mRNAs encoding α-microtubulin, RRM2, and proteins associated with the cell cycle [43]. In virus-induced murine mammary tumors, the eIF3e gene was identified as a common insertion site and suggested that production of truncated eIF3e could lead to malignant transformation of mammary epithelial cells [44]. In prostate cancer, elevation of eIF3h positively correlates with tumor stages. The expression level of eIF3h is higher in metastatic prostate cancer than in primary prostate cancer, and eIF3h may play an important regulatory role in the translation of specific mRNAs [45]. Therefore, aberrant overexpression of eIF3h may contribute to tumor development by upregulating the translation of important mRNAs associated with cell proliferation [46]. Other translation factors are also dysregulated in cancer. eIF1 expression is downregulated in pancreatic ductal adenocarcinoma [47]. eIF1A is essential for cell proliferation and the cell cycle in cancer [48]. Interestingly, although eIF2α phosphorylation leads to reduced global translation, the translation of a restricted subset of mRNAs is enhanced, which facilitates glycolysis and cell invasion in cancer [49,50][49][50]. eIF5 is overexpressed in colorectal cancer and hepatocellular carcinoma and predicts poor prognosis [22,51][22][51]. eIF6 is reported to be markedly upregulated in hepatocellular carcinoma, colorectal cancer, and gallbladder cancer, which lead to tumor progression via mTOR and AKT-related signaling pathways [52,53,54,55][52][53][54][55]. Targeting eIF6-mediated translation blunts lipid accumulation and oncogenic transformation in the liver [56]. eEF1A plays an important and well-defined role in cancer development and progression [26,57][26][57], and eEF1A is aberrantly highly expressed in a variety of tumors and suggests a poor prognosis [58,59][58][59]. eEF2 also plays an important role in promoting the progression of tumors such as breast cancer [38,60,61][38][60][61]. Taken together, these translation factors affect the translation initiation and elongation process of multiple mRNAs in different types of cancer.
As a key enzyme regulating mRNA fate, the regulation of METTL3 on the translation process in tumors cannot be ignored. In recent years, many studies have reported that METTL3 could lead to changes in the expression of target genes through the regulation of mRNA translation process, which in turn caused tumor progression. The importance of RBPs in canonical translation machinery in the aberrant translation of m6A-modified mRNAs was also mentioned in many studies [7,62][7][62]. Therefore, the following is intended to introduce the various mechanisms involved in the translational regulation of mRNA by METTL3 in cancer and the interconnection between various m6A reading proteins and canonical translation machinery in these processes, so as to provide new ideas and possibilities targeting METTL3-mediated translation regulation for cancer treatment.
References
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975.
- Bokar, J.A.; Shambaugh, M.E.; Polayes, D.; Matera, A.G.; Rottman, F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 1997, 3, 1233–1247.
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 2012, 149, 1635–1646.
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206.
- Linder, B.; Grozhik, A.; Olarerin-George, A.; Meydan, C. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772.
- Perry, R.P.; Scherrer, K. The methylated constituents of globin mRNA. FEBS Lett. 1975, 57, 73–78.
- Wang, X.; Zhao, B.S.; Roundtree, I.A. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399.
- Tang, Y.; Chen, K.; Song, B.; Ma, J.; Wu, X.; Xu, Q.; Wei, Z.; Su, J.; Liu, G.; Rong, R.; et al. m6A-Atlas: A comprehensive knowledgebase for unraveling the N6-methyladenosine (m6A) epitranscriptome. Nucleic Acids Res. 2021, 49, D134–D143.
- Geula, S.; Moshitch-Moshkovitz, S.; Dominissini, D.; Mansour, A.A.; Kol, N.; Salmon-Divon, M.; Hershkovitz, V.; Peer, E.; Mor, N.; Manor, Y.S.; et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 2015, 347, 1002–1006.
- Yoon, K.-J.; Ringeling, F.R.; Vissers, C.; Jacob, F.; Pokrass, M.; Jimenez-Cyrus, D.; Su, Y.; Kim, N.-S.; Zhu, Y.; Zheng, L.; et al. Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation. Cell 2017, 171, 877–889.e17.
- Livneh, I.; Moshitch-Moshkovitz, S.; Amariglio, N.; Rechavi, G.; Dominissini, D. The m 6 A epitranscriptome: Transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 2020, 21, 36–51.
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624.
- Deng, L.J.; Deng, W.Q.; Fan, S.R.; Chen, M.F.; Qi, M.; Lyu, W.Y.; Qi, Q.; Tiwari, A.K.; Chen, J.X.; Zhang, D.M.; et al. m6A modification: Recent advances, anticancer targeted drug discovery and beyond. Mol. Cancer 2022, 21, 52.
- Oerum, S.; Meynier, V.; Catala, M.; Tisné, C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021, 49, 7239–7255.
- Cheng, M.; Sheng, L.; Gao, Q.; Xiong, Q.; Zhang, H.; Wu, M.; Liang, Y.; Zhu, F.; Zhang, Y.; Zhang, X.; et al. The m6A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-κB/MYC signaling network. Oncogene 2019, 38, 3667–3680.
- Liu, L.; Wang, J.; Sun, G.; Wu, Q.; Ma, J.; Zhang, X.; Huang, N.; Bian, Z.; Gu, S.; Xu, M.; et al. m6A mRNA methylation regulates CTNNB1 to promote the proliferation of hepatoblastoma. Mol. Cancer 2019, 18, 188.
- Wang, W.; Shao, F.; Yang, X.; Wang, J.; Zhu, R.; Yang, Y.; Zhao, G.; Guo, D.; Sun, Y.; Wang, J.; et al. METTL3 promotes tumour development by decreasing APC expression mediated by APC mRNA N6-methyladenosine-dependent YTHDF binding. Nat. Commun. 2021, 12, 3803.
- Cui, Q.; Shi, H.; Ye, P.; Li, L.; Qu, Q.; Sun, G.; Sun, G.; Lu, Z.; Huang, Y.; Yang, C.-G.; et al. m(6)A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017, 18, 2622–2634.
- Wu, Y.; Chang, N.; Zhang, Y.; Zhang, X.; Xu, L.; Che, Y.; Qiao, T.; Bin Wu, B.; Zhou, Y.; Jiang, J.; et al. METTL3-mediated m(6)A mRNA modification of FBXW7 suppresses lung adenocarcinoma. J. Exp. Clin. Cancer Res. 2021, 40, 90.
- Buttgereit, F.; Brand, M.D. A hierarchy of ATP-consuming processes in mammalian cells. Biochem. J. 1995, 312, 163–167.
- Schwanhäusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342.
- Bhat, M.; Robichaud, N.; Hulea, L.; Sonenberg, N.; Pelletier, J.; Topisirovic, I. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015, 14, 261–278.
- Colina, R.; Costa-Mattioli, M.; Dowling, R.J.O.; Jaramillo, M.; Tai, L.-H.; Breitbach, C.J.; Martineau, Y.; Larsson, O.; Rong, L.; Svitkin, Y.V.; et al. Translational control of the innate immune response through IRF-7. Nature 2008, 452, 323–328.
- Larsson, O.; Perlman, D.M.; Fan, D.; Reilly, C.S.; Peterson, M.; Dahlgren, C.; Liang, Z.; Li, S.; Polunovsky, V.A.; Wahlestedt, C.; et al. Apoptosis resistance downstream of eIF4E: Posttranscriptional activation of an anti-apoptotic transcript carrying a consensus hairpin structure. Nucleic Acids Res. 2006, 34, 4375–4386.
- Ruggero, D. Translational control in cancer etiology. Cold Spring Harb. Perspect. Biol. 2013, 5, a012336.
- Rubio, A.; Garland, G.D.; Sfakianos, A. Aberrant protein synthesis and cancer development: The role of canonical eukaryotic initiation, elongation and termination factors in tumorigenesis. Semin. Cancer Biol. 2022, 86, 151–165.
- De Benedetti, A.; Graff, J.R. eIF-4E expression and its role in malignancies and metastases. Oncogene 2004, 23, 3189–3199.
- Feoktistova, K.; Tuvshintogs, E.; Do, A.; Fraser, C.S. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc. Natl. Acad. Sci. USA 2013, 110, 13339–13344.
- Merrick, W.C.; Pavitt, G.D. Protein Synthesis Initiation in Eukaryotic Cells. Cold Spring Harb. Perspect. Biol. 2018, 10, a033092.
- Kozak, M. New ways of initiating translation in eukaryotes? Mol. Cell Biol. 2001, 21, 1899–1907.
- Bhardwaj, U.; Powell, P.; Goss, D.J. Eukaryotic initiation factor (eIF) 3 mediates Barley Yellow Dwarf Viral mRNA 3’-5’ UTR interactions and 40S ribosomal subunit binding to facilitate cap-independent translation. Nucleic Acids Res. 2019, 47, 6225–6235.
- Hinnebusch, A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014, 83, 779–812.
- Rios-Fuller, T.J.; Mahe, M.; Walters, B.; Abbadi, D.; Pérez-Baos, S. Translation Regulation by eIF2α Phosphorylation and mTORC1 Signaling Pathways in Non-Communicable Diseases (NCDs). Int. J. Mol. Sci. 2020, 21, 5301.
- Sanvito, F.; Piatti, S.; Villa, A.; Bossi, M.; Lucchini, G.; Marchisio, P.C.; Biffo, S. The beta4 integrin interactor p27(BBP/eIF6) is an essential nuclear matrix protein involved in 60S ribosomal subunit assembly. J. Cell Biol. 1999, 144, 823–837.
- Basu, U.; Si, K.; Warner, J.R.; Maitra, U. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol. Cell Biol. 2001, 21, 1453–1462.
- Gandin, V.; Miluzio, A.; Barbieri, A.M.; Beugnet, A.; Kiyokawa, H.; Marchisio, P.C.; Biffo, S. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature 2008, 455, 684–688.
- Miluzio, A.; Beugnet, A.; Volta, V.; Biffo, S. Eukaryotic initiation factor 6 mediates a continuum between 60S ribosome biogenesis and translation. EMBO Rep. 2009, 10, 459–465.
- Proud, C.G. Regulation and roles of elongation factor 2 kinase. Biochem. Soc. Trans. 2015, 43, 328–332.
- Robichaud, N.; Sonenberg, N.; Ruggero, D.; Schneider, R.J. Translational Control in Cancer. Cold Spring Harb. Perspect. Biol. 2019, 11, a032896.
- Kevil, C.G.; De Benedetti, A.; Payne, D.K.; Coe, L.L.; Laroux, F.S.; Alexander, J.S. Translational regulation of vascular permeability factor by eukaryotic initiation factor 4E: Implications for tumor angiogenesis. Int. J. Cancer 1996, 65, 785–790.
- Nathan, C.-A.; Carter, P.; Liu, L.; Li, B.D.; Abreo, F.; Tudor, A.; Zimmer, S.G.; De Benedetti, A. Elevated expression of eIF4E and FGF-2 isoforms during vascularization of breast carcinomas. Oncogene 1997, 15, 1087–1094.
- Bachmann, F.; Bänziger, R.; Burger, M.M. Cloning of a novel protein overexpressed in human mammary carcinoma. Cancer Res. 1997, 57, 988–994.
- Dong, Z.; Zhang, J.T. EIF3 p170, a mediator of mimosine effect on protein synthesis and cell cycle progression. Mol. Biol. Cell 2003, 14, 3942–3951.
- Mayeur, G.L.; Hershey, J.W. Malignant transformation by the eukaryotic translation initiation factor 3 subunit p48 (eIF3e). FEBS Lett. 2002, 514, 49–54.
- Nupponen, N.N.; Porkka, K.; Kakkola, L.; Tanner, M.; Persson, K.; Borg, A.; Isola, J.; Visakorpi, T. Amplification and overexpression of p40 subunit of eukaryotic translation initiation factor 3 in breast and prostate cancer. Am. J. Pathol. 1999, 154, 1777–1783.
- Saramäki, O.; Willi, N.; Bratt, O.; Gasser, T.C.; Koivisto, P.; Nupponen, N.N.; Bubendorf, L.; Visakorpi, T. Amplification of EIF3S3 gene is associated with advanced stage in prostate cancer. Am. J. Pathol. 2001, 159, 2089–2094.
- Golob-Schwarzl, N.; Puchas, P.; Gogg-Kamerer, M.; Weichert, W.; Göppert, B.; Haybaeck, J. New Pancreatic Cancer Biomarkers eIF1, eIF2D, eIF3C and eIF6 Play a Major Role in Translational Control in Ductal Adenocarcinoma. Anticancer Res. 2020, 40, 3109–3118.
- Sehrawat, U.; Koning, F.; Ashkenazi, S.; Stelzer, G.; Leshkowitz, D.; Dikstein, R. Cancer-Associated Eukaryotic Translation Initiation Factor 1A Mutants Impair Rps3 and Rps10 Binding and Enhance Scanning of Cell Cycle Genes. Mol. Cell Biol. 2019, 39, e00441-18.
- García-Jiménez, C.; Goding, C.R. Starvation and Pseudo-Starvation as Drivers of Cancer Metastasis through Translation Reprogramming. Cell Metab. 2019, 29, 254–267.
- Li, Z.; Ge, Y.; Dong, J.; Wang, H.; Zhao, T.; Wang, X.; Liu, J.; Gao, S.; Shi, L.; Yang, S.; et al. BZW1 Facilitates Glycolysis and Promotes Tumor Growth in Pancreatic Ductal Adenocarcinoma Through Potentiating eIF2α Phosphorylation. Gastroenterology 2022, 162, 1256–1271.e14.
- Golob-Schwarzl, N.; Krassnig, S.; Toeglhofer, A.M.; Park, Y.N.; Gogg-Kamerer, M.; Vierlinger, K.; Schröder, F.; Rhee, H.; Schicho, R.; Fickert, P.; et al. New liver cancer biomarkers: PI3K/AKT/mTOR pathway members and eukaryotic translation initiation factors. Eur. J. Cancer 2017, 83, 56–70.
- Sun, L.; Liu, S.; Wang, X.; Zheng, X.; Chen, Y.; Shen, H. eIF6 promotes the malignant progression of human hepatocellular carcinoma via the mTOR signaling pathway. J. Transl. Med. 2021, 19, 216.
- Scagliola, A.; Miluzio, A.; Mori, G.; Ricciardi, S.; Oliveto, S.; Manfrini, N.; Biffo, S. Inhibition of eIF6 Activity Reduces Hepatocellular Carcinoma Growth: An In Vivo and In Vitro Study. Int. J. Mol. Sci. 2022, 23, 7720.
- Lin, J.; Yu, X.; Xie, L.; Wang, P.; Li, T.; Xiao, Y.; Zhou, J.; Peng, S.; Huang, J.; Luo, Y.; et al. eIF6 Promotes Colorectal Cancer Proliferation and Invasion by Regulating AKT-Related Signaling Pathways. J. Biomed. Nanotechnol. 2019, 15, 1556–1567.
- Golob-Schwarzl, N.; Wodlej, C.; Kleinegger, F.; Gogg-Kamerer, M.; Birkl-Toeglhofer, A.M.; Petzold, J.; Aigelsreiter, A.; Thalhammer, M.; Park, Y.N.; Haybaeck, J. Eukaryotic translation initiation factor 6 overexpression plays a major role in the translational control of gallbladder cancer. J. Cancer Res. Clin. Oncol. 2019, 145, 2699–2711.
- Scagliola, A.; Miluzio, A.; Ventura, G.; Oliveto, S.; Cordiglieri, C. Targeting of eIF6-driven translation induces a metabolic rewiring that reduces NAFLD and the consequent evolution to hepatocellular carcinoma. Nat. Commun. 2021, 12, 4878.
- Abbas, W.; Kumar, A.; Herbein, G. The eEF1A Proteins: At the Crossroads of Oncogenesis, Apoptosis, and Viral Infections. Front. Oncol. 2015, 7, 75.
- Lee, S.; Francoeur, A.; Liu, E.W.S. Tissue-specific expression in mammalian brain, heart, and muscle of S1, a member of the elongation factor-1 alpha gene family. J. Biol. Chem. 1992, 267, 24064–24068.
- Tomlinson, V.A.; Newbery, H.J.; Wray, N.R.; Jackson, J.; Larionov, A.; Miller, W.R.; Dixon, J.M. Translation elongation factor eEF1A2 is a potential oncoprotein that is overexpressed in two-thirds of breast tumours. BMC Cancer 2005, 12, 113.
- Liu, R.; Proud, C.G. Eukaryotic elongation factor 2 kinase as a drug target in cancer, and in cardiovascular and neurodegenerative diseases. Acta Pharmacol. Sin. 2016, 37, 285–294.
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 8, 1393–1406.
- Choe, J.; Lin, S.; Zhang, W.; Liu, Q.; Wang, L.; Ramirez-Moya, J.; Du, P.; Kim, W.; Tang, S.; Sliz, P.; et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 2018, 561, 556–560.
More
