You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Magdalena Oroń.
Alternative pre-mRNA splicing is a process that allows for the generation of an extremely diverse proteome from a much smaller number of genes. In this process, non-coding introns are excised from primary mRNA and coding exons are joined together. Different combinations of exons give rise to alternative versions of a protein.
- alternative splicing
- mutant p53
- KRAS
- CMYC
- splicing factors
1. Introduction
Alternative pre-mRNA splicing is a common process in higher eukaryotes that allows for the generation of an extremely diverse proteome from a much smaller number of genes [1,2][1][2]. It is estimated that 95% of human multiexon genes undergo alternative splicing [3]. Alternative splicing plays a crucial role in development and tissue differentiation by providing proteins with different functions from the same set of genes [4]. Disruption of this process may result in disturbed cell differentiation and oncogenesis. The pattern of alternative exon selection in cancer resembles the one in corresponding embryonic tissues and involves the same regulatory pathways [5]. Multiple mRNA-Seq datasets comparing tumors and corresponding normal tissues demonstrate that an aberrant splicing landscape is a common feature of cancer [6,7,8][6][7][8]. Alternative oncogenic isoforms of different proteins are involved in all hallmarks of cancer: growth signal independence, avoidance of apoptosis, unlimited proliferation, genome instability, motility and invasiveness, angiogenesis, and metabolism [9,10][9][10].
mRNA splicing involves the spliceosome (a large ribonucleoprotein complex composed of snRNAs and associated proteins) and splicing regulatory factors [11]. In hematological malignancies, point mutations in spliceosome components (e.g., SF3B1, SF3A1, U2AF1, or ZRSR2) are more frequent, while in solid tumors, splicing is deregulated mostly by upregulation or downregulation of splicing regulatory factors (e.g., SRSF1, TRA2β, hnRNPA1, or QKI) [12]. Splicing is tightly regulated at various levels: by linking with transcription rate [13,14][13][14], expression, nonsense-mediated decay, and post-translational modifications of splicing factors [15,16,17][15][16][17]. In addition, relative levels of snRNAs U1, U2, U4, U5, and U6—basic components of the spliceosome—may also be important [18]. All of these can be disrupted by the activation of proto-oncogenes.
Altered splicing may also increase the activity of driver oncogenes and oncogenic signaling pathways via preferential expression of alternative pro-oncogenic isoforms, creation of new splice sites leading to the expression of cancer-specific isoforms, or indirectly via changed splicing of regulatory proteins. Specific examples will be discussed in the following paragraphs.
2. Alternative Splicing under the Control of Driver Oncogenes
2.1. CMYC
The oncogenic activity of CMYC in different cancers has been extensively studied and well documented. This proto-oncogene belongs to the family of transcription factors, along with NMYC and LMYC [23][19]. In normal tissues, its level is low and tightly regulated on the transcriptional and post-transcriptional level. In cancers, CMYC level is frequently increased via various mechanisms: gene amplification or translocation, upregulation of signaling pathways boosting CMYC activity, post-translational modifications, and increased protein stability [24,25][20][21]. Activated CMYC is a hallmark of many cancers and is required for their initiation and maintenance [26][22]. The transcriptional program of CMYC and its intersections with other oncogenes have been broadly discussed elsewhere [27,28][23][24]. Here, wresearchers will focus only on the role of CMYC in the regulation of alternative splicing and its consequences for oncogenesis. The globally increased transcription induced by CMYC requires adaptation of the splicing and translation machinery. The shortage of spliceosome components and splicing regulatory factors affects splicing profile [29,30][25][26]. CMYC hyperactivation increases translation of a core spliceosome component, SF3A3, through an eIF3D-dependent mechanism. This affects the splicing of many mRNAs, including those involved in mitochondrial metabolism favoring stem-cell-like, cancer-associated changes [31][27]. Koh et al. demonstrated that CMYC directly upregulates the transcription of several small nuclear ribonucleoprotein particle assembly genes, including PRMT5, an arginine methyltransferase. This protein methylates Sm proteins that form the Sm core of the spliceosomal snRNPs U1, U2, U4/U6, and U5, and is critical for correct spliceosome assembly [32][28]. The perturbation in PRMT5 expression caused by CMYC leads mainly to intron retention or exon skipping and disturbs splicing fidelity [30][26]. CMYC not only causes overexpression of the most studied oncogenic splicing factor, SRSF1 [33][29], but also causes overexpression of several other proteins from the same family. Urbanski et al. identified an alternative splicing (AS) signature associated with high CMYC activity in breast cancer and showed that the change in AS is caused by the co-expression of splicing factors modules. They also demonstrated that overexpression of at least one module of the splicing factors SRSF2, SRSF3, and SRSF7 correlated with high CMYC activity across thirty-three cancer types [34][30]. The transcriptional activity of CMYC indirectly affects other oncogenes and key oncogenic processes by deregulating splicing. Through the overexpression of hnRNPH splicing factor, CMYC affects the RAS/RAF/ERK signaling pathway. A high level of CMYC and hnRNPH correlates with the expression of full-length A-RAF kinase. A-RAF activates oncogenic RAS signaling and inhibits apoptosis by binding to proapoptotic MST2 kinase. In cells with a low level of CMYC and hnRNPH, A-RAF is spliced into a shorter isoform that cannot bind to MST2. A-RAFshort still binds to RAS, but in a way that inhibits RAS activation [35][31]. One of the hallmarks of cancer is the conversion of glucose into energy via aerobic glycolysis instead of oxidative phosphorylation. This process is governed by PKM2, a pyruvate kinase isoform expressed mainly in embryonic and cancer cells, in contrast to the PKM1 isoform present in most adult normal cells. Overactive CMYC upregulates transcription of PTBP1, hnRNPA1, and hnRNPA2 splicing factors, resulting in a high PKM2/PKM1 ratio [36][32]. Zhang et al. found that another member of the MYC family, NMYC, similarly causes the upregulation of the same splicing factors, PTBP1 and HNRNPA1, leading to the expression of pro-oncogenic PMK2 isoform [37][33]. CMYC not only initiates transcription but also regulates transcription rate [38][34]. A pre-mRNA splicing occurs co-transcriptionally and the transcription rate influences the selection of alternative exons [13,39][13][35]. CMYC controls the transcription and splicing of a splicing factor, Sam68 pre-mRNA, in prostate cancer by binding to the promoter and increasing the RNA Pol II processivity [40][36]. Splicing factors (SF) frequently use an autoregulatory loop to control their own level: when SF protein level is high, it induces incorporation of a poison exon that triggers nonsense-mediated decay (NMD) of the transcript. In prostate and breast cancer, it was shown that CMYC not only increased the transcription of SRSF3 (and several other splicing factors) but also prevented incorporation of the poison exon [41][37]. The exact mechanism remains to be elucidated, but one may hypothesize that the exon skipping is caused by the increased transcription rate induced by CMYC. Selected effects of activated CMYC on splicing are summarized in Figure 1.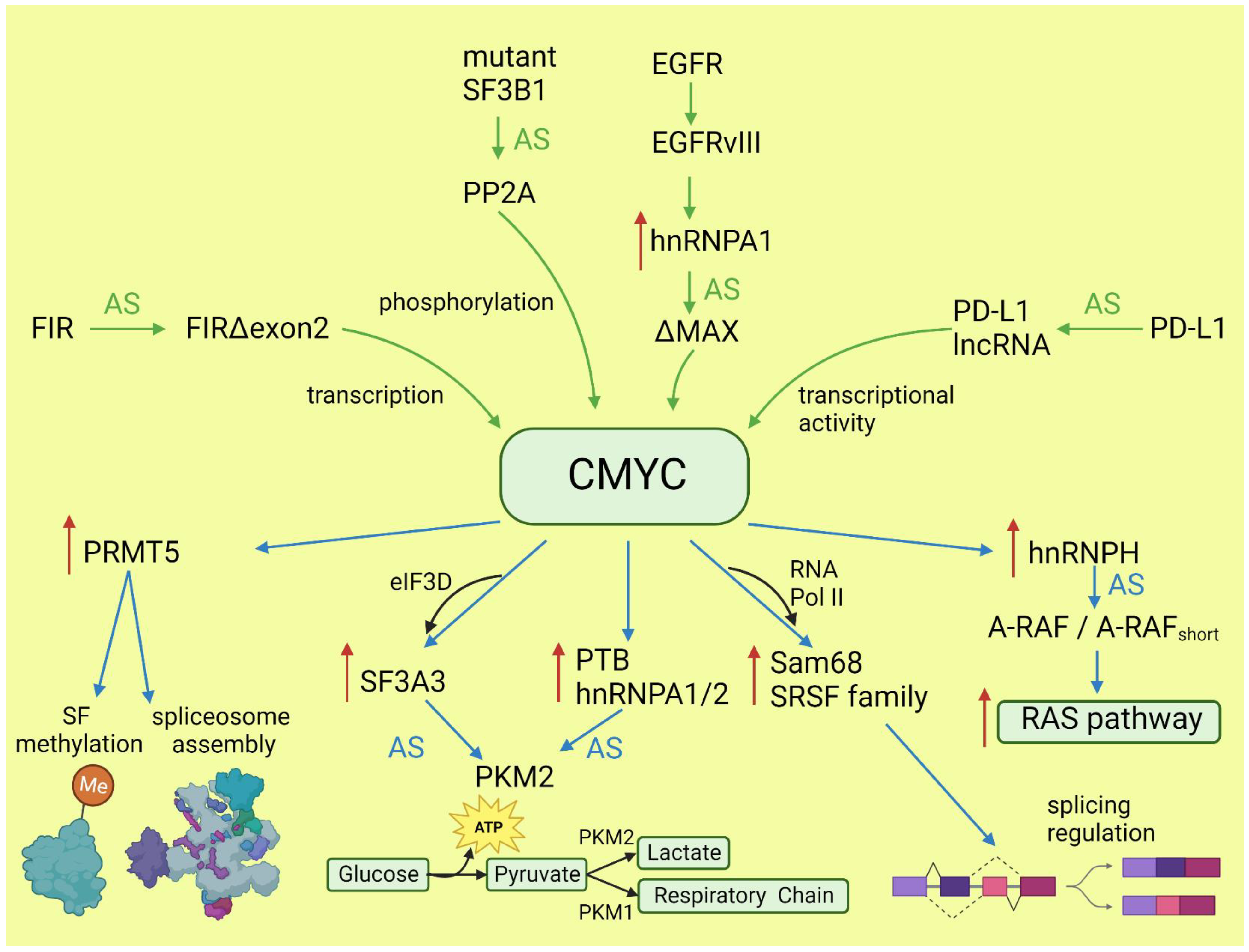
Figure 1. Effects of CMYC on splicing (blue arrows) and the influence of altered splicing on CMYC (green arrows). Red arrows: increased expression; AS: alternative splicing; SF: splicing factor.
2.2. RAS and Downstream Pathways
KRAS, together with NRAS and HRAS, belongs to the small GTPases family, which under physiological conditions cycles between active (GTP-bound) and inactive (GDP-bound) conformations in response to stimulation of cell surface receptors such as HER2, EGFR, or CMET. KRAS is commonly mutated in human cancers; however, the percentage of mutations varies between cancer types [42][38]. Mutations most frequently occur in codons for glycine at amino acid position 12 or 13, causing KRAS hyperactivity that contributes to its transforming properties [43][39]. Signaling cascades are triggered by the binding of the active RAS to RAS-binding domains within several known RAS effector pathways. The most important, in the cancer context, are the PI3K-AKT and MAPK signaling pathways [44][40]. Lo et al. found that in lung cancer cells expressing either WT or mutant KRAS variants, the phosphorylation of SR splicing factors was reduced in those with mutant KRAS expression. This observation correlated with changed cassette exon skipping or inclusion in mutant vs. WT KRAS [45][41]. Correct phosphorylation of SR proteins by SRPK1 and CLK1 kinases is indispensable for their binding to splicing enhancer sequences on pre-mRNA [46][42]. It is known that signaling through PI3K/AKT activates SRPK1 kinase [47][43]; however, the link between RAS and SRPK1 activation was not directly demonstrated. In hepatocellular carcinoma, activation of the RAS/PI3K/AKT pathway leads to the expression of KLF6 splicing variant 1 (KLF6-SV1), which antagonizes the function of full-length KLF6 [48][44]. This protein is a tumor suppressor. The sorter isoform is an oncogene found in many tumors and involved in proliferation, metastasis, and angiogenesis [49][45]. KLF6 splicing is regulated by SRSF1 splicing factor [50][46]. Cheng et al. found a positive feedback loop involving activated RAS and an alternative isoform of CD44 (CD44v6) that can be a coreceptor of growth factor receptors. Activation of the RAS/MAPK/ERK pathway correlated with a preference for exon v6 inclusion. CD44v6, in turn, sustained signaling through tyrosine receptor kinases which activate RAS and downstream pathways, boosting cancer cell proliferation. The author suggested that this positive feedback loop could be a mechanism of the RAS-dependent pathway’s activation in cancers without oncogenic RAS mutations [51][47]. Alternative splicing of CD44 is regulated by a splicing factor, Sam68, which is phosphorylated by ERK, an effector of the RAS/MAPK/ERK pathway [52][48]. The same signaling pathway may be responsible for alternative pro-oncogenic splicing of other Sam68-dependent genes, namely, CCND1, SRSF1, BCL-xL, and mTOR [53][49]. Notably, mTOR is a key effector part of the PI3K/AKT/mTOR pathway that can be activated by RAS [44][40]. The effects of mutated KRAS on splicing are summarized in Figure 2.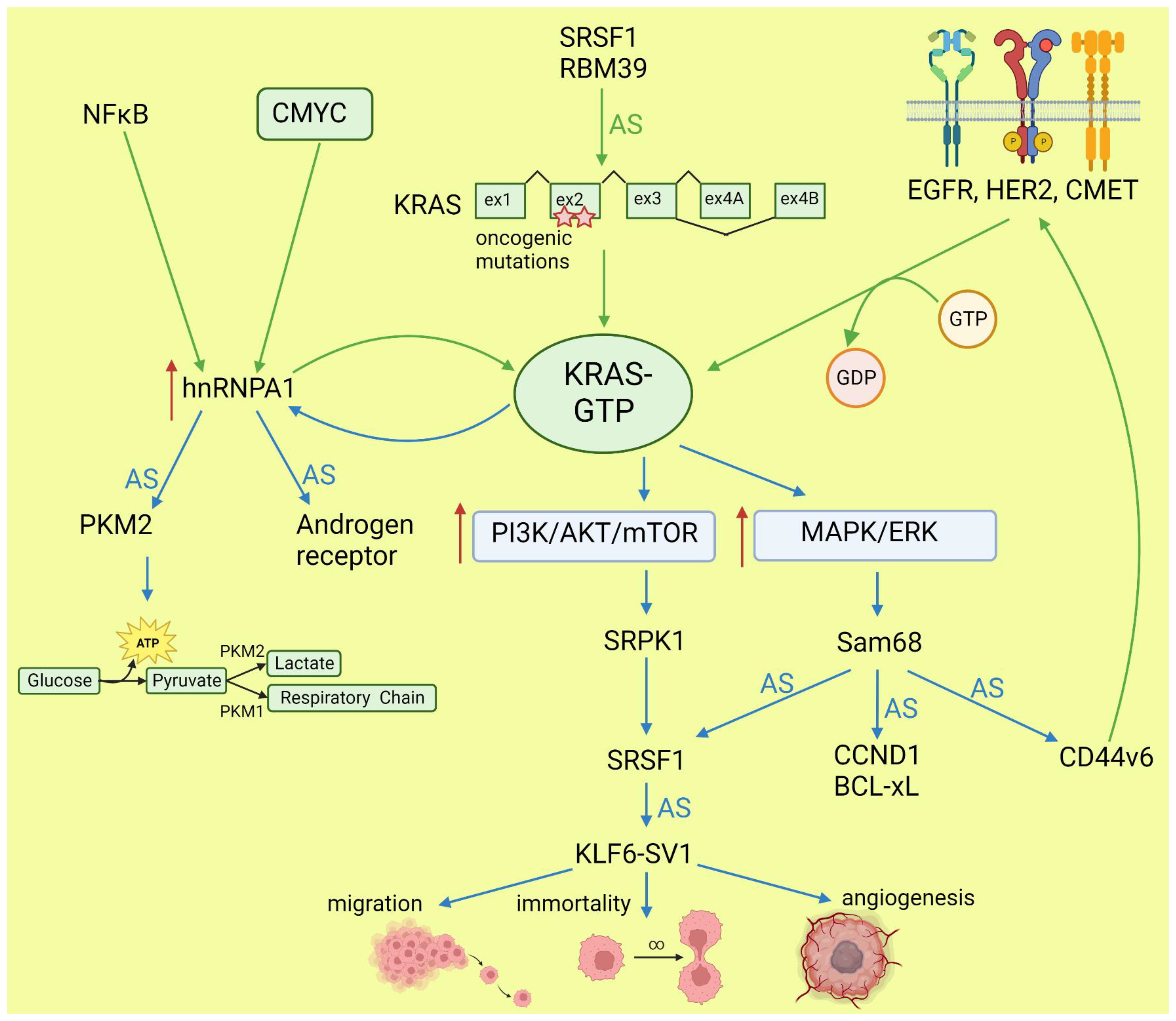
Figure 2. Effects of KRAS on splicing (blue arrows) and influence of altered splicing on KRAS (green arrows). AS: alternative splicing; red arrows: increased expression or pathway activity.
2.3. Mutant p53
The TP53 gene is a tumor suppressor, while its hotspot mutants gain oncogenic activity (named gain of function, GOF) and are involved in all hallmarks of cancer [54][50]. In contrast to wild-type proteins, p53 GOF mutants do not bind directly to promoters, but interact with different transcription factors to control expression of target genes [55][51]. In addition to transcription control, p53 mutants contribute to oncogenesis via interaction with TAp63 and TAp73 [56][52] to inhibit their proapoptotic activity, and with ID4 in angiogenesis stimulation [57,58][53][54]. While many researchers have studied the alternative splicing of the TP53 transcript and demonstrated the role of alternative isoforms in cancer development (discussed in more detail later), little is known about the role of mutant p53 in alternative splicing regulation. Escobar-Hoyos et al. demonstrated that in pancreatic cancer, mutant p53 upregulates the expression of hnRNPK splicing factor. This leads to the mis-splicing of GAP proteins and activates the oncogenic RAS pathway [59][55]. In addition, the RNA-Seq results underlying this study indicate other genes alternatively spliced under the influence of the p53 mutant. Pruszko et al. found, in breast cancer, a ribonucleoprotein complex composed of mutant p53, a splicing factor SRSF1, ID4, and lncRNA MALAT1. The complex altered a splicing of the VEGFA transcript, thus promoting proangiogenic isoforms over antiangiogenic isoforms [58][54]. A proangiogenic role of the studied complex was confirmed in vivo in a zebrafish model [60][56]. These two independent studies indicated that GOF p53 mutants influence alternative splicing in cancer. However, published results are limited to two cancer types and focused on a few target genes. Further studies based on a broad spectrum of tumors are needed to understand the role and interactions with other oncogenes of the GOF p53 mutants in alternative splicing regulation.3. Driver Oncogenes Regulated by Alternative Splicing
3.1. TP53 Regulation by Splicing
The TP53 gene produces at least 12 isoforms with different features that might prevent or promote cancer development. The canonical, full-length p53 (FLp53) protein has seven functional domains: N-terminal transactivation domains TAD1 and TAD2, a proline-rich domain, a DNA-binding domain, a nuclear localization signal, an oligomerization domain, and a negative-regulation domain [61][57]. The α (full-length), β, and γ variants of p53 result from alternative splicing of the C-terminus. β and γ isoforms are formed by partial retention of intron 9 (i9). The resulting alternative exons, 9b or 9g, contain a stop codon, leading to the replacement of the oligomerization domain into 10 amino acids (DQTSFQKENC) in p53β and 15 amino acids (MLLDLRWCYFLINSS) in p53γ (Figure 3). SRSF1 and SRSF3 regulate the splicing of i9 and favor the expression of FLp53 [62,63][58][59]. Marcel et al. observed that inhibiting CLK kinase or silencing SRSF1 upregulated the expression of p53β and p53γ isoforms in the breast cancer cell line MCF7 [63][59]. In contrast to SRSF1 and SRSF3, SRSF7 has been reported to enhance p53β expression in response to ionizing radiation [64][60]. Each of the α, β, and γ isoforms may also be changed from the N-terminus through alternative splicing of intron 2 (Δ40), alternative start of translation from internal IRES (Δ40, Δ160), or transcription from internal promoter (Δ133, Δ160) [65,66][61][62] (Figure 3). The co-expression of so many isoforms and the difficulty of distinguishing between them using available molecular biology methods make it challenging to evaluate their role in cancer.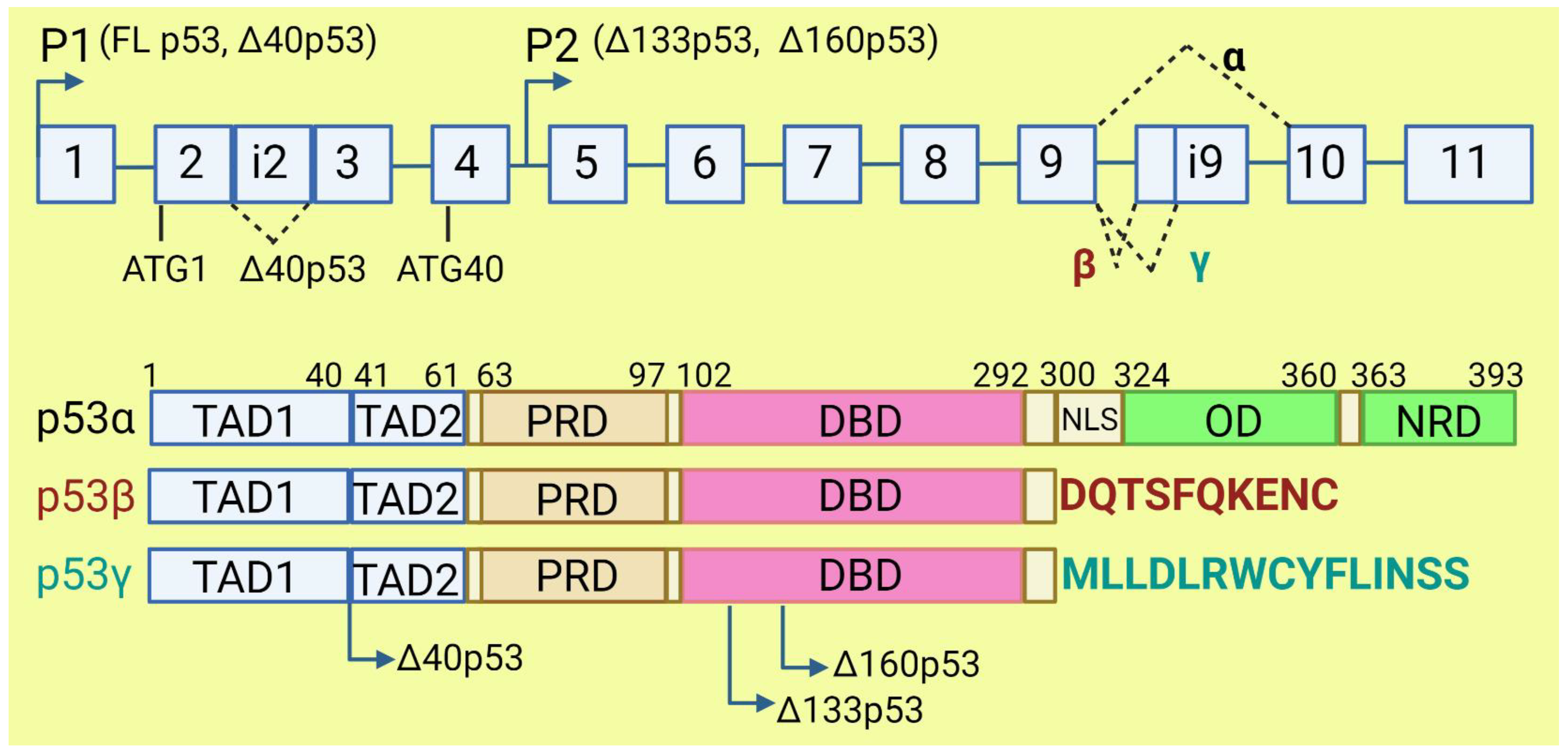
Figure 3. The human TP53 gene and p53 isoforms. (Upper panel): schematic representation of TP53 gene. Exons are numbered from 1 to 11. Alternative splicing of introns i2 and i9 provides alternative p53 isoforms. P1 and P2: alternative promoters. ATG1 and ATG40: alternative transcription start sites. (Lower panel): representation of p53 domains. TAD: transactivation domain; PRD: proline rich domain; DBD: DNA binding domain; NLS: nuclear localization signal; OD: oligomerization domain; NRD: negative regulation domain. Isoforms β and γ lack C-terminal domains, including OD, replaced by DQTSFQKENC and MLLDLRWCYFLINSS, respectively. Isoforms Δ40p53, Δ133p53, and Δ160p53 have a deletion from N-terminus.
3.2. CMYC
Functional alternative isoforms of CMYC are not reported in the literature but alternative splicing of other proteins may affect CMYC activity in cancers. FUSE-binding protein-interacting repressor (FIR) is a suppressor of CMYC transcription. A splice variant of FIR that lacks exon 2 in the transcriptional repressor domain (FIRΔexon2) presents dominant negative activity towards full-length FIR; thus, the expression of FIRΔexon2 upregulates CMYC transcription. The increased ratio of FIRΔexon2/FIR contributes to human colorectal and hepatocellular carcinomas as well as lymphomas [104][100]. Mutations in SF3B1, the gene coding the most commonly mutated spliceosome component across cancers, alters the splicing and promotes the decay of mRNA, coding a specific subunit of the PP2A serine/threonine phosphatase and increasing phosphorylation and, consequently, the stability of CMYC [105][101]. This observation is particularly valuable, because SF3B1 may be targeted with several inhibitors, and it was demonstrated that patients with overactive MYC are more sensitive to these drugs [29][25]. In lung adenocarcinoma, under the influence of IFNγ, the protein coding gene PD-L1 may be spliced into long non-coding RNA PL-L1-lnc. This alternative transcript does not encode a protein but is functional. Through direct interaction with CMYC, it boosts its activity as a transcription factor and contributes to the increased proliferation and invasiveness of neoplastic cells [106][102]. EGFRvIII is a constitutively active variant of EGFR formed by genomic rearrangement. It is highly expressed in glioblastoma multiforme (GBM), where is detected at an overall frequency of 25–64% [107][103]. EGFRvIII induces a broad change in the alternative splicing program via upregulation of the heterogeneous nuclear ribonucleoprotein A1 splicing factor. HnRNPA1 promotes inclusion of exon 5 in the transcript encoding the CMYC-interacting partner MAX. The resulting shorter isoform ΔMAX enhances CMYC-dependent transformation [108][104]. The impact of alternative splicing on CMYC is summarized in Figure 1.3.3. KRAS and Downstream Pathways
KRAS may be spliced into two isoforms with alternative exon 4: KRAS4A and KRAS4B. Both isoforms are oncogenic if they have a mutation in G12 or G13 [109][105]. The two isoforms differ in their C-terminal domain, referred to as the hypervariable region (HVR), which determines the possible post-translational modifications and membrane localization [110][106]. Although most of the interactome is the same for both isoforms, some proteins interact specifically with one or the other [111,112][107][108]. This is related to the distinct association to the membrane and intracellular localization. For example, KRAS4A undergoes palmitoylation–depalmitoylation cycles, unlike KRAS4B. This allows it to bind to heterokinase HK1 on the outer mitochondrial membrane and stimulate HK1 activity, thereby increasing glycolysis in cancer cells [113][109]. Moreover, KRAS4A binds more strongly to RAF1, augmenting downstream ERK signaling and anchorage-independent growth [112][108]. The regulation of KRAS splicing in cancers is not clear. Hall et al. reported that in colon cancer with inactive APC, the key role is played by SRSF1 [114][110]. KRAS4A expression may be also decreased by splicing inhibitor Indisulfam, which targets the protein RBM39 associated with the spliceosome [115][111] (Figure 2). Alternative splicing also affects downstream components of RAS/MAPK and PI3K/AKT pathways [116][112]. A-RAF, B-RAF, and C-RAF can be spliced into dominant-negative isoforms with only an RAS-binding domain or constitutively active, oncogenic isoforms containing only a kinase domain. The splicing of A-RAF in cancer is regulated by hnRNPH, and indirectly by CMYC, which upregulates the level of the splicing factor [35][31]. In hepatocellular carcinoma, it was demonstrated that hnRNPA2 also causes constitutive activation of the RAS pathway by the splicing of A-RAF [117][113]. The MKNK2 gene is expressed as two isoforms: MNK2a and MNK2b kinases. The alternative splicing of MKNK2 is regulated by proto-oncogene SRSF1 [118][114]. While MNK2a can suppress RAS-induced transformation, MNK2b is pro-oncogenic. It phosphorylates the translation initiating factor eIF4E—an effector of RAS-dependent pathways—increasing translation in cancers [119][115]. In the PI3K-AKT-mTOR pathway, both PI3K and mTOR may be spliced into more active oncogenic isoforms [120,121][116][117]. The mTORβ isoform, despite losing most of the protein–protein interaction domains, retains its kinase activity and ability to complex with Rictor and Raptor proteins. Most importantly, mTORβ accelerates proliferation by shortening the G1 phase of the cell cycle, and its overexpression is sufficient for immortal cell transformation [121][117]. Increased activation of AKT further affects aberrant alternative splicing by phosphorylation of SRPK kinase [47][43].References
- Matlin, A.J.; Clark, F.; Smith, C.W. Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell. Biol. 2005, 6, 386–398.
- Blencowe, B.J. Alternative splicing: New insights from global analyses. Cell 2006, 126, 37–47.
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415.
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell. Biol. 2017, 18, 437–451.
- Singh, A.; Rajeevan, A.; Gopalan, V.; Agrawal, P.; Day, C.P.; Hannenhalli, S. Broad misappropriation of developmental splicing profile by cancer in multiple organs. Nat. Commun. 2022, 13, 7664.
- Xiong, W.; Gao, D.; Li, Y.; Liu, X.; Dai, P.; Qin, J.; Wang, G.; Li, K.; Bai, H.; Li, W. Genome-wide profiling of chemoradiationinduced changes in alternative splicing in colon cancer cells. Oncol. Rep. 2016, 36, 2142–2150.
- Yang, C.; Wu, Q.; Huang, K.; Wang, X.; Yu, T.; Liao, X.; Huang, J.; Zhu, G.; Gong, Y.; Han, C.; et al. Genome-Wide Profiling Reveals the Landscape of Prognostic Alternative Splicing Signatures in Pancreatic Ductal Adenocarcinoma. Front. Oncol. 2019, 9, 511.
- Danan-Gotthold, M.; Golan-Gerstl, R.; Eisenberg, E.; Meir, K.; Karni, R.; Levanon, E.Y. Identification of recurrent regulated alternative splicing events across human solid tumors. Nucleic Acids Res. 2015, 43, 5130–5144.
- Oltean, S.; Bates, D.O. Hallmarks of alternative splicing in cancer. Oncogene 2014, 33, 5311–5318.
- Ladomery, M. Aberrant alternative splicing is another hallmark of cancer. Int. J. Cell Biol. 2013, 2013, 463786.
- Anczukow, O.; Krainer, A.R. Splicing-factor alterations in cancers. RNA 2016, 22, 1285–1301.
- Urbanski, L.M.; Leclair, N.; Anczuków, O. Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip. Rev. RNA 2018, 9, e1476.
- de la Mata, M.; Alonso, C.R.; Kadener, S.; Fededa, J.P.; Blaustein, M.; Pelisch, F.; Cramer, P.; Bentley, D.; Kornblihtt, A.R. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 2003, 12, 525–532.
- Shukla, S.; Oberdoerffer, S. Co-transcriptional regulation of alternative pre-mRNA splicing. Biochim. Biophys. Acta 2012, 1819, 673–683.
- Lareau, L.F.; Inada, M.; Green, R.E.; Wengrod, J.C.; Brenner, S.E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 2007, 446, 926–929.
- Corkery, D.P.; Holly, A.C.; Lahsaee, S.; Dellaire, G. Connecting the speckles: Splicing kinases and their role in tumorigenesis and treatment response. Nucleus 2015, 6, 279–288.
- Aubol, B.E.; Plocinik, R.M.; Hagopian, J.C.; Ma, C.T.; McGlone, M.L.; Bandyopadhyay, R.; Fu, X.D.; Adams, J.A. Partitioning RS domain phosphorylation in an SR protein through the CLK and SRPK protein kinases. J. Mol. Biol. 2013, 425, 2894–2909.
- Dvinge, H.; Guenthoer, J.; Porter, P.L.; Bradley, R.K. RNA components of the spliceosome regulate tissue- and cancer-specific alternative splicing. Genome Res. 2019, 29, 1591–1604.
- Kress, T.R.; Sabo, A.; Amati, B. MYC: Connecting selective transcriptional control to global RNA production. Nat. Rev. Cancer 2015, 15, 593–607.
- Spencer, C.A.; Groudine, M. Control of c-myc regulation in normal and neoplastic cells. Adv. Cancer Res. 1991, 56, 1–48.
- Kalkat, M.; De Melo, J.; Hickman, K.A.; Lourenco, C.; Redel, C.; Resetca, D.; Tamachi, A.; Tu, W.B.; Penn, L.Z. MYC Deregulation in Primary Human Cancers. Genes 2017, 8, 151.
- Gabay, M.; Li, Y.; Felsher, D.W. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 2014, 4, a014241.
- Grzes, M.; Oron, M.; Staszczak, Z.; Jaiswar, A.; Nowak-Niezgoda, M.; Walerych, D. A Driver Never Works Alone-Interplay Networks of Mutant p53, MYC, RAS, and Other Universal Oncogenic Drivers in Human Cancer. Cancers 2020, 12, 1532.
- Van Dang, C.; McMahon, S.B. Emerging Concepts in the Analysis of Transcriptional Targets of the MYC Oncoprotein: Are the Targets Targetable? Genes Cancer 2010, 1, 560–567.
- Hsu, T.Y.; Simon, L.M.; Neill, N.J.; Marcotte, R.; Sayad, A.; Bland, C.S.; Echeverria, G.V.; Sun, T.; Kurley, S.J.; Tyagi, S.; et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 2015, 525, 384–388.
- Koh, C.M.; Bezzi, M.; Low, D.H.; Ang, W.X.; Teo, S.X.; Gay, F.P.; Al-Haddawi, M.; Tan, S.Y.; Osato, M.; Sabo, A.; et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 2015, 523, 96–100.
- Ciesla, M.; Ngoc, P.C.T.; Cordero, E.; Martinez, A.S.; Morsing, M.; Muthukumar, S.; Beneventi, G.; Madej, M.; Munita, R.; Jonsson, T.; et al. Oncogenic translation directs spliceosome dynamics revealing an integral role for SF3A3 in breast cancer. Mol. Cell 2021, 81, 1453–1468.
- Meister, G.; Eggert, C.; Buhler, D.; Brahms, H.; Kambach, C.; Fischer, U. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol. 2001, 11, 1990–1994.
- Das, S.; Anczukow, O.; Akerman, M.; Krainer, A.R. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 2012, 1, 110–117.
- Urbanski, L.; Brugiolo, M.; Park, S.; Angarola, B.L.; Leclair, N.K.; Yurieva, M.; Palmer, P.; Sahu, S.K.; Anczukow, O. MYC regulates a pan-cancer network of co-expressed oncogenic splicing factors. Cell Rep. 2022, 41, 111704.
- Rauch, J.; Moran-Jones, K.; Albrecht, V.; Schwarzl, T.; Hunter, K.; Gires, O.; Kolch, W. c-Myc regulates RNA splicing of the A-Raf kinase and its activation of the ERK pathway. Cancer Res. 2011, 71, 4664–4674.
- David, C.J.; Chen, M.; Assanah, M.; Canoll, P.; Manley, J.L. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 2010, 463, 364–368.
- Zhang, S.; Wei, J.S.; Li, S.Q.; Badgett, T.C.; Song, Y.K.; Agarwal, S.; Coarfa, C.; Tolman, C.; Hurd, L.; Liao, H.; et al. MYCN controls an alternative RNA splicing program in high-risk metastatic neuroblastoma. Cancer Lett. 2016, 371, 214–224.
- Baluapuri, A.; Hofstetter, J.; Dudvarski Stankovic, N.; Endres, T.; Bhandare, P.; Vos, S.M.; Adhikari, B.; Schwarz, J.D.; Narain, A.; Vogt, M.; et al. MYC Recruits SPT5 to RNA Polymerase II to Promote Processive Transcription Elongation. Mol. Cell 2019, 74, 674–687.
- Girard, C.; Will, C.L.; Peng, J.; Makarov, E.M.; Kastner, B.; Lemm, I.; Urlaub, H.; Hartmuth, K.; Luhrmann, R. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat. Commun. 2012, 3, 994.
- Caggiano, C.; Pieraccioli, M.; Panzeri, V.; Sette, C.; Bielli, P. c-MYC empowers transcription and productive splicing of the oncogenic splicing factor Sam68 in cancer. Nucleic Acids Res. 2019, 47, 6160–6171.
- Phillips, J.W.; Pan, Y.; Tsai, B.L.; Xie, Z.; Demirdjian, L.; Xiao, W.; Yang, H.T.; Zhang, Y.; Lin, C.H.; Cheng, D.; et al. Pathway-guided analysis identifies Myc-dependent alternative pre-mRNA splicing in aggressive prostate cancers. Proc. Natl. Acad. Sci. USA 2020, 117, 5269–5279.
- Prior, I.A.; Hood, F.E.; Hartley, J.L. The Frequency of Ras Mutations in Cancer. Cancer Res. 2020, 80, 2969–2974.
- Prior, I.A.; Lewis, P.D.; Mattos, C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012, 72, 2457–2467.
- Gimple, R.C.; Wang, X. RAS: Striking at the Core of the Oncogenic Circuitry. Front. Oncol. 2019, 9, 965.
- Lo, A.; McSharry, M.; Berger, A.H. Oncogenic KRAS alters splicing factor phosphorylation and alternative splicing in lung cancer. BMC Cancer 2022, 22, 1315.
- Naro, C.; Sette, C. Phosphorylation-mediated regulation of alternative splicing in cancer. Int. J. Cell Biol. 2013, 2013, 151839.
- Zhou, Z.; Qiu, J.; Liu, W.; Zhou, Y.; Plocinik, R.M.; Li, H.; Hu, Q.; Ghosh, G.; Adams, J.A.; Rosenfeld, M.G.; et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol. Cell 2012, 47, 422–433.
- Yea, S.; Narla, G.; Zhao, X.; Garg, R.; Tal-Kremer, S.; Hod, E.; Villanueva, A.; Loke, J.; Tarocchi, M.; Akita, K.; et al. Ras promotes growth by alternative splicing-mediated inactivation of the KLF6 tumor suppressor in hepatocellular carcinoma. Gastroenterology 2008, 134, 1521–1531.
- Hu, K.; Zheng, Q.K.; Ma, R.J.; Ma, C.; Sun, Z.G.; Zhang, N. Kruppel-Like Factor 6 Splice Variant 1: An Oncogenic Transcription Factor Involved in the Progression of Multiple Malignant Tumors. Front. Cell Dev. Biol. 2021, 9, 661731.
- Munoz, U.; Puche, J.E.; Hannivoort, R.; Lang, U.E.; Cohen-Naftaly, M.; Friedman, S.L. Hepatocyte growth factor enhances alternative splicing of the Kruppel-like factor 6 (KLF6) tumor suppressor to promote growth through SRSF1. Mol. Cancer Res. 2012, 10, 1216–1227.
- Cheng, C.; Yaffe, M.B.; Sharp, P.A. A positive feedback loop couples Ras activation and CD44 alternative splicing. Genes Dev. 2006, 20, 1715–1720.
- Matter, N.; Herrlich, P.; Konig, H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 2002, 420, 691–695.
- Frisone, P.; Pradella, D.; Di Matteo, A.; Belloni, E.; Ghigna, C.; Paronetto, M.P. SAM68: Signal Transduction and RNA Metabolism in Human Cancer. BioMed Res. Int. 2015, 2015, 528954.
- Walerych, D.; Napoli, M.; Collavin, L.; Del Sal, G. The rebel angel: Mutant p53 as the driving oncogene in breast cancer. Carcinogenesis 2012, 33, 2007–2017.
- Oren, M.; Rotter, V. Mutant p53 gain-of-function in cancer. Cold Spring Harb. Perspect. Biol. 2010, 2, a001107.
- Schilling, T.; Kairat, A.; Melino, G.; Krammer, P.H.; Stremmel, W.; Oren, M.; Muller, M. Interference with the p53 family network contributes to the gain of oncogenic function of mutant p53 in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2010, 394, 817–823.
- Fontemaggi, G.; Dell’Orso, S.; Trisciuoglio, D.; Shay, T.; Melucci, E.; Fazi, F.; Terrenato, I.; Mottolese, M.; Muti, P.; Domany, E.; et al. The execution of the transcriptional axis mutant p53, E2F1 and ID4 promotes tumor neo-angiogenesis. Nat. Struct. Mol. Biol. 2009, 16, 1086–1093.
- Pruszko, M.; Milano, E.; Forcato, M.; Donzelli, S.; Ganci, F.; Di Agostino, S.; De Panfilis, S.; Fazi, F.; Bates, D.O.; Bicciato, S.; et al. The mutant p53-ID4 complex controls VEGFA isoforms by recruiting lncRNA MALAT1. EMBO Rep. 2017, 18, 1331–1351.
- Escobar-Hoyos, L.F.; Penson, A.; Kannan, R.; Cho, H.; Pan, C.H.; Singh, R.K.; Apken, L.H.; Hobbs, G.A.; Luo, R.; Lecomte, N.; et al. Altered RNA Splicing by Mutant p53 Activates Oncogenic RAS Signaling in Pancreatic Cancer. Cancer Cell 2020, 38, 198–211.
- Pruszko, M.; Milano, E.; Zylicz, A.; Zylicz, M.; Blandino, G.; Fontemaggi, G. Zebrafish as experimental model to establish the contribution of mutant p53 and ID4 to breast cancer angiogenesis in vivo. J. Thorac. Dis. 2018, 10, E231–E233.
- Surget, S.; Khoury, M.P.; Bourdon, J.C. Uncovering the role of p53 splice variants in human malignancy: A clinical perspective. OncoTargets Ther. 2013, 7, 57–68.
- Tang, Y.; Horikawa, I.; Ajiro, M.; Robles, A.I.; Fujita, K.; Mondal, A.M.; Stauffer, J.K.; Zheng, Z.M.; Harris, C.C. Downregulation of splicing factor SRSF3 induces p53beta, an alternatively spliced isoform of p53 that promotes cellular senescence. Oncogene 2013, 32, 2792–2798.
- Marcel, V.; Fernandes, K.; Terrier, O.; Lane, D.P.; Bourdon, J.C. Modulation of p53beta and p53gamma expression by regulating the alternative splicing of TP53 gene modifies cellular response. Cell Death Differ. 2014, 21, 1377–1387.
- Chen, J.; Crutchley, J.; Zhang, D.; Owzar, K.; Kastan, M.B. Identification of a DNA Damage-Induced Alternative Splicing Pathway That Regulates p53 and Cellular Senescence Markers. Cancer Discov. 2017, 7, 766–781.
- Steffens Reinhardt, L.; Zhang, X.; Wawruszak, A.; Groen, K.; De Iuliis, G.N.; Avery-Kiejda, K.A. Good Cop, Bad Cop: Defining the Roles of Delta40p53 in Cancer and Aging. Cancers 2020, 12, 1659.
- Joruiz, S.M.; Beck, J.A.; Horikawa, I.; Harris, C.C. The Delta133p53 Isoforms, Tuners of the p53 Pathway. Cancers 2020, 12, 3422.
- Solomon, H.; Sharon, M.; Rotter, V. Modulation of alternative splicing contributes to cancer development: Focusing on p53 isoforms, p53beta and p53gamma. Cell Death Differ. 2014, 21, 1347–1349.
- Bourdon, J.C.; Khoury, M.P.; Diot, A.; Baker, L.; Fernandes, K.; Aoubala, M.; Quinlan, P.; Purdie, C.A.; Jordan, L.B.; Prats, A.C.; et al. p53 mutant breast cancer patients expressing p53gamma have as good a prognosis as wild-type p53 breast cancer patients. Breast Cancer Res. 2011, 13, R7.
- Avery-Kiejda, K.A.; Morten, B.; Wong-Brown, M.W.; Mathe, A.; Scott, R.J. The relative mRNA expression of p53 isoforms in breast cancer is associated with clinical features and outcome. Carcinogenesis 2014, 35, 586–596.
- Oh, L.; Hainaut, P.; Blanchet, S.; Ariffin, H. Expression of p53 N-terminal isoforms in B-cell precursor acute lymphoblastic leukemia and its correlation with clinicopathological profiles. BMC Cancer 2020, 20, 110.
- Hofstetter, G.; Berger, A.; Berger, R.; Zoric, A.; Braicu, E.I.; Reimer, D.; Fiegl, H.; Marth, C.; Zeimet, A.G.; Ulmer, H.; et al. The N-terminally truncated p53 isoform Delta40p53 influences prognosis in mucinous ovarian cancer. Int. J. Gynecol. Cancer 2012, 22, 372–379.
- Avery-Kiejda, K.A.; Zhang, X.D.; Adams, L.J.; Scott, R.J.; Vojtesek, B.; Lane, D.P.; Hersey, P. Small molecular weight variants of p53 are expressed in human melanoma cells and are induced by the DNA-damaging agent cisplatin. Clin. Cancer Res. 2008, 14, 1659–1668.
- Hafsi, H.; Santos-Silva, D.; Courtois-Cox, S.; Hainaut, P. Effects of Delta40p53, an isoform of p53 lacking the N-terminus, on transactivation capacity of the tumor suppressor protein p53. BMC Cancer 2013, 13, 134.
- Kitayner, M.; Rozenberg, H.; Kessler, N.; Rabinovich, D.; Shaulov, L.; Haran, T.E.; Shakked, Z. Structural basis of DNA recognition by p53 tetramers. Mol. Cell 2006, 22, 741–753.
- Sun, Y.; Manceau, A.; Frydman, L.; Cappuccio, L.; Neves, D.; Basso, V.; Wang, H.; Fombonne, J.; Maisse, C.; Mehlen, P.; et al. Delta40p53 isoform up-regulates netrin-1/UNC5B expression and potentiates netrin-1 pro-oncogenic activity. Proc. Natl. Acad. Sci. USA 2021, 118, e2103319118.
- Ungewitter, E.; Scrable, H. Delta40p53 controls the switch from pluripotency to differentiation by regulating IGF signaling in ESCs. Genes Dev. 2010, 24, 2408–2419.
- Gong, L.; Gong, H.; Pan, X.; Chang, C.; Ou, Z.; Ye, S.; Yin, L.; Yang, L.; Tao, T.; Zhang, Z.; et al. p53 isoform Delta113p53/Delta133p53 promotes DNA double-strand break repair to protect cell from death and senescence in response to DNA damage. Cell Res. 2015, 25, 351–369.
- Kazantseva, M.; Mehta, S.; Eiholzer, R.A.; Gimenez, G.; Bowie, S.; Campbell, H.; Reily-Bell, A.L.; Roth, I.; Ray, S.; Drummond, C.J.; et al. The Delta133p53beta isoform promotes an immunosuppressive environment leading to aggressive prostate cancer. Cell Death Dis. 2019, 10, 631.
- Ozretic, P.; Hanzic, N.; Proust, B.; Sabol, M.; Trnski, D.; Radic, M.; Musani, V.; Ciribilli, Y.; Milas, I.; Puljiz, Z.; et al. Expression profiles of p53/p73, NME and GLI families in metastatic melanoma tissue and cell lines. Sci. Rep. 2019, 9, 12470.
- Fujita, K.; Mondal, A.M.; Horikawa, I.; Nguyen, G.H.; Kumamoto, K.; Sohn, J.J.; Bowman, E.D.; Mathe, E.A.; Schetter, A.J.; Pine, S.R.; et al. p53 isoforms Delta133p53 and p53beta are endogenous regulators of replicative cellular senescence. Nat. Cell Biol. 2009, 11, 1135–1142.
- Kazantseva, M.; Eiholzer, R.A.; Mehta, S.; Taha, A.; Bowie, S.; Roth, I.; Zhou, J.; Joruiz, S.M.; Royds, J.A.; Hung, N.A.; et al. Elevation of the TP53 isoform Delta133p53beta in glioblastomas: An alternative to mutant p53 in promoting tumor development. J. Pathol. 2018, 246, 77–88.
- Gadea, G.; Arsic, N.; Fernandes, K.; Diot, A.; Joruiz, S.M.; Abdallah, S.; Meuray, V.; Vinot, S.; Anguille, C.; Remenyi, J.; et al. TP53 drives invasion through expression of its Delta133p53beta variant. Elife 2016, 5, e14734.
- Campbell, H.; Fleming, N.; Roth, I.; Mehta, S.; Wiles, A.; Williams, G.; Vennin, C.; Arsic, N.; Parkin, A.; Pajic, M.; et al. ∆133p53 isoform promotes tumour invasion and metastasis via interleukin-6 activation of JAK-STAT and RhoA-ROCK signalling. Nat. Commun. 2018, 9, 254.
- Bernard, H.; Garmy-Susini, B.; Ainaoui, N.; Van Den Berghe, L.; Peurichard, A.; Javerzat, S.; Bikfalvi, A.; Lane, D.P.; Bourdon, J.C.; Prats, A.C. The p53 isoform, Delta133p53alpha, stimulates angiogenesis and tumour progression. Oncogene 2013, 32, 2150–2160.
- Aoubala, M.; Murray-Zmijewski, F.; Khoury, M.P.; Fernandes, K.; Perrier, S.; Bernard, H.; Prats, A.C.; Lane, D.P.; Bourdon, J.C. p53 directly transactivates Delta133p53alpha, regulating cell fate outcome in response to DNA damage. Cell Death Differ. 2011, 18, 248–258.
- Candeias, M.M.; Hagiwara, M.; Matsuda, M. Cancer-specific mutations in p53 induce the translation of Delta160p53 promoting tumorigenesis. EMBO Rep. 2016, 17, 1542–1551.
- Marcel, V.; Perrier, S.; Aoubala, M.; Ageorges, S.; Groves, M.J.; Diot, A.; Fernandes, K.; Tauro, S.; Bourdon, J.C. Delta160p53 is a novel N-terminal p53 isoform encoded by Delta133p53 transcript. FEBS Lett. 2010, 584, 4463–4468.
- Tadijan, A.; Precazzini, F.; Hanzic, N.; Radic, M.; Gavioli, N.; Vlasic, I.; Ozretic, P.; Pinto, L.; Skreblin, L.; Barban, G.; et al. Altered Expression of Shorter p53 Family Isoforms Can Impact Melanoma Aggressiveness. Cancers 2021, 13, 5231.
- Lai, M.Y.; Chang, H.C.; Li, H.P.; Ku, C.K.; Chen, P.J.; Sheu, J.C.; Huang, G.T.; Lee, P.H.; Chen, D.S. Splicing mutations of the p53 gene in human hepatocellular carcinoma. Cancer Res. 1993, 53, 1653–1656.
- Austin, F.; Oyarbide, U.; Massey, G.; Grimes, M.; Corey, S.J. Synonymous mutation in TP53 results in a cryptic splice site affecting its DNA-binding site in an adolescent with two primary sarcomas. Pediatr. Blood Cancer 2017, 64, e26584.
- Eicheler, W.; Zips, D.; Dorfler, A.; Grenman, R.; Baumann, M. Splicing mutations in TP53 in human squamous cell carcinoma lines influence immunohistochemical detection. J. Histochem. Cytochem. 2002, 50, 197–204.
- Smeby, J.; Sveen, A.; Eilertsen, I.A.; Danielsen, S.A.; Hoff, A.M.; Eide, P.W.; Johannessen, B.; Hektoen, M.; Skotheim, R.I.; Guren, M.G.; et al. Transcriptional and functional consequences of TP53 splice mutations in colorectal cancer. Oncogenesis 2019, 8, 35.
- Holmila, R.; Fouquet, C.; Cadranel, J.; Zalcman, G.; Soussi, T. Splice mutations in the p53 gene: Case report and review of the literature. Hum. Mutat. 2003, 21, 101–102.
- Bartel, F.; Pinkert, D.; Fiedler, W.; Kappler, M.; Wurl, P.; Schmidt, H.; Taubert, H. Expression of alternatively and aberrantly spliced transcripts of the MDM2 mRNA is not tumor-specific. Int. J. Oncol. 2004, 24, 143–151.
- Inoue, K.; Fry, E.A. Aberrant splicing of the DMP1-ARF-MDM2-p53 pathway in cancer. Int. J. Cancer 2016, 139, 33–41.
- Lukashchuk, N.; Vousden, K.H. Ubiquitination and degradation of mutant p53. Mol. Cell. Biol. 2007, 27, 8284–8295.
- Traweek, R.S.; Cope, B.M.; Roland, C.L.; Keung, E.Z.; Nassif, E.F.; Erstad, D.J. Targeting the MDM2-p53 pathway in dedifferentiated liposarcoma. Front. Oncol. 2022, 12, 1006959.
- Momand, J.; Jung, D.; Wilczynski, S.; Niland, J. The MDM2 gene amplification database. Nucleic Acids Res. 1998, 26, 3453–3459.
- Zheng, T.; Wang, J.; Zhao, Y.; Zhang, C.; Lin, M.; Wang, X.; Yu, H.; Liu, L.; Feng, Z.; Hu, W. Spliced MDM2 isoforms promote mutant p53 accumulation and gain-of-function in tumorigenesis. Nat. Commun. 2013, 4, 2996.
- Wiech, M.; Olszewski, M.B.; Tracz-Gaszewska, Z.; Wawrzynow, B.; Zylicz, M.; Zylicz, A. Molecular mechanism of mutant p53 stabilization: The role of HSP70 and MDM2. PLoS ONE 2012, 7, e51426.
- Comiskey, D.F., Jr.; Montes, M.; Khurshid, S.; Singh, R.K.; Chandler, D.S. SRSF2 Regulation of MDM2 Reveals Splicing as a Therapeutic Vulnerability of the p53 Pathway. Mol. Cancer Res. 2020, 18, 194–203.
- Comiskey, D.F., Jr.; Jacob, A.G.; Singh, R.K.; Tapia-Santos, A.S.; Chandler, D.S. Splicing factor SRSF1 negatively regulates alternative splicing of MDM2 under damage. Nucleic Acids Res. 2015, 43, 4202–4218.
- Aptullahoglu, E.; Ciardullo, C.; Wallis, J.P.; Marr, H.; Marshall, S.; Bown, N.; Willmore, E.; Lunec, J. Splicing Modulation Results in Aberrant Isoforms and Protein Products of p53 Pathway Genes and the Sensitization of B Cells to Non-Genotoxic MDM2 Inhibition. Int. J. Mol. Sci. 2023, 24, 2410.
- Matsushita, K.; Tomonaga, T.; Shimada, H.; Shioya, A.; Higashi, M.; Matsubara, H.; Harigaya, K.; Nomura, F.; Libutti, D.; Levens, D.; et al. An essential role of alternative splicing of c-myc suppressor FUSE-binding protein-interacting repressor in carcinogenesis. Cancer Res. 2006, 66, 1409–1417.
- Liu, Z.; Yoshimi, A.; Wang, J.; Cho, H.; Chun-Wei Lee, S.; Ki, M.; Bitner, L.; Chu, T.; Shah, H.; Liu, B.; et al. Mutations in the RNA Splicing Factor SF3B1 Promote Tumorigenesis through MYC Stabilization. Cancer Discov. 2020, 10, 806–821.
- Qu, S.; Jiao, Z.; Lu, G.; Yao, B.; Wang, T.; Rong, W.; Xu, J.; Fan, T.; Sun, X.; Yang, R.; et al. PD-L1 lncRNA splice isoform promotes lung adenocarcinoma progression via enhancing c-Myc activity. Genome Biol. 2021, 22, 104.
- Heimberger, A.B.; Suki, D.; Yang, D.; Shi, W.; Aldape, K. The natural history of EGFR and EGFRvIII in glioblastoma patients. J. Transl. Med. 2005, 3, 38.
- Babic, I.; Anderson, E.S.; Tanaka, K.; Guo, D.; Masui, K.; Li, B.; Zhu, S.; Gu, Y.; Villa, G.R.; Akhavan, D.; et al. EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 2013, 17, 1000–1008.
- Voice, J.K.; Klemke, R.L.; Le, A.; Jackson, J.H. Four human ras homologs differ in their abilities to activate Raf-1, induce transformation, and stimulate cell motility. J. Biol. Chem. 1999, 274, 17164–17170.
- Nuevo-Tapioles, C.; Philips, M.R. The role of KRAS splice variants in cancer biology. Front. Cell Dev. Biol. 2022, 10, 1033348.
- Miller, S.P.; Maio, G.; Zhang, X.; Badillo Soto, F.S.; Zhu, J.; Ramirez, S.Z.; Lin, H. A Proteomic Approach Identifies Isoform-Specific and Nucleotide-Dependent RAS Interactions. Mol. Cell. Proteom. 2022, 21, 100268.
- Zhang, X.; Cao, J.; Miller, S.P.; Jing, H.; Lin, H. Comparative Nucleotide-Dependent Interactome Analysis Reveals Shared and Differential Properties of KRas4a and KRas4b. ACS Cent. Sci. 2018, 4, 71–80.
- Amendola, C.R.; Mahaffey, J.P.; Parker, S.J.; Ahearn, I.M.; Chen, W.C.; Zhou, M.; Court, H.; Shi, J.; Mendoza, S.L.; Morten, M.J.; et al. KRAS4A directly regulates hexokinase 1. Nature 2019, 576, 482–486.
- Hall, A.E.; Pohl, S.O.; Cammareri, P.; Aitken, S.; Younger, N.T.; Raponi, M.; Billard, C.V.; Carrancio, A.B.; Bastem, A.; Freile, P.; et al. RNA splicing is a key mediator of tumour cell plasticity and a therapeutic vulnerability in colorectal cancer. Nat. Commun. 2022, 13, 2791.
- Chen, W.C.; To, M.D.; Westcott, P.M.K.; Delrosario, R.; Kim, I.J.; Philips, M.; Tran, Q.; Bollam, S.R.; Goodarzi, H.; Bayani, N.; et al. Targeting KRAS4A splicing through the RBM39/DCAF15 pathway inhibits cancer stem cells. Nat. Commun. 2021, 12, 4288.
- Siegfried, Z.; Bonomi, S.; Ghigna, C.; Karni, R. Regulation of the Ras-MAPK and PI3K-mTOR Signalling Pathways by Alternative Splicing in Cancer. Int. J. Cell Biol. 2013, 2013, 568931.
- Shilo, A.; Ben Hur, V.; Denichenko, P.; Stein, I.; Pikarsky, E.; Rauch, J.; Kolch, W.; Zender, L.; Karni, R. Splicing factor hnRNP A2 activates the Ras-MAPK-ERK pathway by controlling A-Raf splicing in hepatocellular carcinoma development. RNA 2014, 20, 505–515.
- Mogilevsky, M.; Shimshon, O.; Kumar, S.; Mogilevsky, A.; Keshet, E.; Yavin, E.; Heyd, F.; Karni, R. Modulation of MKNK2 alternative splicing by splice-switching oligonucleotides as a novel approach for glioblastoma treatment. Nucleic Acids Res. 2018, 46, 11396–11404.
- Maimon, A.; Mogilevsky, M.; Shilo, A.; Golan-Gerstl, R.; Obiedat, A.; Ben-Hur, V.; Lebenthal-Loinger, I.; Stein, I.; Reich, R.; Beenstock, J.; et al. Mnk2 alternative splicing modulates the p38-MAPK pathway and impacts Ras-induced transformation. Cell Rep. 2014, 7, 501–513.
- Thorpe, L.M.; Yuzugullu, H.; Zhao, J.J. PI3K in cancer: Divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer 2015, 15, 7–24.
- Panasyuk, G.; Nemazanyy, I.; Zhyvoloup, A.; Filonenko, V.; Davies, D.; Robson, M.; Pedley, R.B.; Waterfield, M.; Gout, I. mTORbeta splicing isoform promotes cell proliferation and tumorigenesis. J. Biol. Chem. 2009, 284, 30807–30814.
More
