This article is associated with the MDPI International Journal of Molecular Sciences article, which belongs to a Special Issue Plant Cell and Organism Development, “Completing Autophagy: Formation and Degradation of the Autophagic Body and Metabolite Salvage in Plants” by Szymon Stefaniak1, Łukasz Wojtyla1, Małgorzata Pietrowska-Borek2, Sławomir Borek1,*
1Department of Plant Physiology, Faculty of Biology, Adam Mickiewicz University Poznań, Uniwersytetu Poznańskiego 6, 61-614 Poznań, Poland; szymon.stefaniak@amu.edu.pl (S.S.); wojtylal@amu.edu.pl (Ł.W.); borek@amu.edu.pl (S.B.)
2Department of Biochemistry and Biotechnology, Faculty of Agronomy and Bioengineering, Poznań University of Life Sciences, Dojazd 11, 60-632 Poznań, Poland; malgorzata.pietrowska-borek@up.poznan.pl (M.P.-B.)
*Correspondence: borek@amu.edu.pl (S.B.)
The article has been published on https://doi.org/10.3390/ijms21062205, Int. J. Mol. Sci. 2020, 21(6), 2205. The work was financed by the National Science Centre, Poland (grant no. 2016/23/B/NZ3/00735).
- Atg proteins
- autophagosome
- tonoplast
- vacuole
- SNARE proteins
1. Introduction
INTRODUCTION
Autophagy, which literally means “self-eating”, plays a crucial role in the degradation of useless or damaged cell components such as macromolecules, protein complexes, and organelles. Autophagy also plays a role in the degradation of foreign elements for cells, such as bacteria, viruses or sperm residues after egg cell fertilization. In plants, autophagy participates in the circulation of cell components and acts as a quality control mechanism. It also functions in some developmental processes such as pollen maturation, aging, and cell death, including programmed cell death [[1][2][3][4][5][6]1,2,3,4,5,6].
2. Formation and trafficking of autophagosome
FORMATION AND TRAFFICKING OF AUTOPHAGOSOME
The first visible symptom of macroautophagy is the appearance in the cytoplasm of a cup-shaped structure, called the phagophore (Figure 1). The phagophore elongates, surrounding and simultaneously separating the fragment of the cytoplasm together with organelles or other components of the cell that are intended for degradation. The final stage of phagophore differentiation is the complete surrounding of the cargo and its sequestration inside the autophagosome. This is a vesicle with a double, bilayer lipid-protein membrane, containing cargo intended for autophagic degradation [[7][8][9][10][11][12]7,8,9,10,11,12]. In plants, the autophagosome fuses with the vacuole creating an autophagic body that is quickly degraded by vacuolar lytic enzymes [[13][14]13,14].
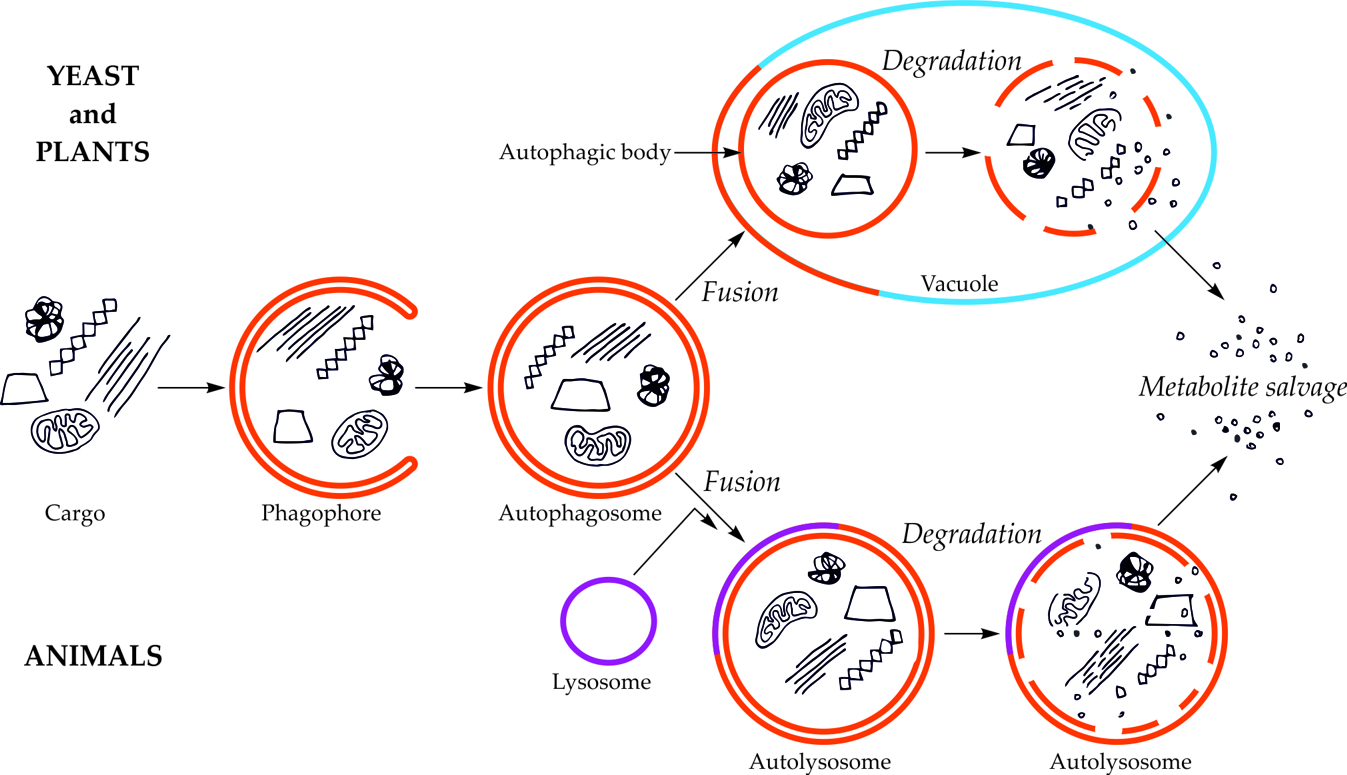
Figure 1. Schematic diagram of macroautophagy in cells of yeast and plants (upper part of the drawing) and in cells of animals (bottom part of the drawing). In yeast and plant cells, the autophagosome fuses to the tonoplast, creating the autophagic body inside the vacuole. In animal cells, the autophagosome fuses with the lysosome, giving the autolysosome. The autophagic body inside the vacuole and the content of autolysosome are rapidly degraded, allowing reuse of metabolites [[15]15].
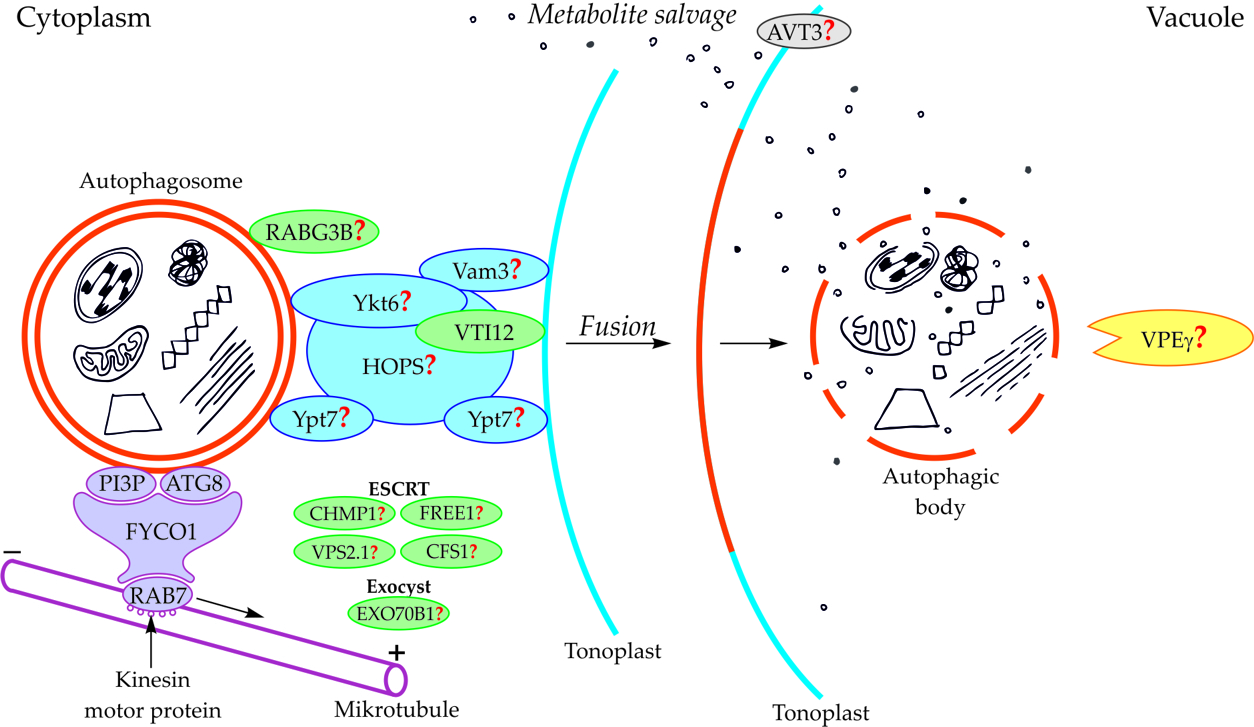
Components of the cytoskeleton play an important role in the cytoplasmic transport of autophagosomes [[16]16]. It is also suggested that the cytoplasmic transport of autophagosomes is enabled by the microtubule network controlled by endosomal sorting complexes required for transport (ESCRT) [[8]8,[17]17,[18]18]. An important role in this transport is played by fully developed autophagosomes with the outer-membrane-anchored Atg8 and phosphatidylinositol 3-phosphate (PI3P) in plants (Figure 2, Table 1) [[19][20][21]19,20,21]. The protein FYCO1 (FYVE and coiled-coil domain-containing protein 1) is also important in the association of the autophagosome membrane and microtubules, and it has a modular structure composed of four amino acid domains and spiral signaling domain FYVE [[20]20,[22]22]. Due to its structure, FYCO1 interacts with the autophagosome surface simultaneously in two places—with Atg8 and PI3P. These two sites for recognition and linking of the autophagosome with the FYCO1 make it possible to distinguish between mature autophagosomes and phagophores [[20]20,23,[23][24]24] because only on the surface of the mature autophagosomes are there simultaneously proteins necessary to form stable and double bonds with FYCO1. Attached to the surface of the autophagosome, FYCO1 also binds to GTP-binding protein 7 (Ypt7) in yeast and Ras-related protein RAB7 (RAB7) in plants [[22]22,25,[25][26]26], creating an autophagosome-FYCO1-Ypt7/RAB7 system that allows the binding of the autophagosome to microtubules. The autophagosome-FYCO1-Ypt7/RAB7 system moves to the plus end of microtubules by the binding of Ypt7/RAB7 to the kinesin motor proteins [[25]25,[27]27].
3. Fusion of the autophagosome with the vacuole and formation of the autophagic body
FUSION OF THE AUTOPHAGOSOME WITH THE VACUOLE AND FORMATION OF THE AUTOPHAGIC BODY
In plants, the fusion of autophagosome and vacuole and the mechanisms regulating this process are poorly understood. So far, only the involvement of protein VTI12 has been confirmed in the fusion of autophagosome and vacuole in plants (Figure 2, Table 1). This protein belongs to the complex named soluble N-ethylmaleimide-sensitive factor activating protein receptors (SNARE proteins). In Arabidopsis mutants with T-DNA VTI12 insertion, growing in rich-nutrient conditions, presented a normal phenotype, whereas under nutrient-poor conditions accelerated aging was observed, confirming that VTI12 is involved in autophagy in plants [[8]8,28,29,30,[28][29][30][31]31]. VTI12 is the only SNARE protein that has been proven to be involved in the fusion of the autophagosome and vacuole in plants (Figure 2, Table 1). Other protein that may be involved in the fusion of autophagosome and vacuole in plants is RABG3B (Figure 2, Table 1). So far, the occurrence of RABG3B has been confirmed in Arabidopsis and Populus and it participates in, among other processes, the formation of the wood conductive elements when programmed cell death occurs [[32][33]32,33]. The protein RABG3B is located at the surface of the autophagosome, however, it remains unclear whether RABG3B can regulate the fusion of the autophagosome and vacuole in plants. It is suggested that the homologous yeast proteins such as Ykt6, Vam3, Ypt7, and complex HOPS are involved in the fusion of autophagosome and vacuole in plants [[20]20,[34]34]. Moreover, it is also suggested that the plant components of the ESCRT complex, such as the charged multi-vesicular body protein 1 (CHMP1), FYVE-domain protein required for endosomal sorting 1 (FREE1), vacuolar protein sorting 2.1 (VPS2.1), cell death-related endosomal FYVE/SYLF protein 1 (CFS1), and plant exocyst complex component EXO70B1 (EXO70B1) are involved in trafficking of the autophagosome, the fusion of autophagosome and vacuole, and the release of the autophagic body into the vacuole [[8]8].
Figure 2. Schematic diagram depicting trafficking and fusion of the autophagosome to the vacuole and the degradation of the autophagic body inside the vacuole in plants. PI3P and ATG8 anchored in the outer membrane of the autophagosome are involved in autophagosome trafficking and bind autophagosome with FYCO1 protein. The complex autophagosome-FYCO1-RAB7 moves along microtubules in the direction of the plus end by the binding of RAB7 to kinesin motor proteins. Protein VTI12 is involved in the fusion of the autophagosome and vacuole. RABG3B is located on the surface of the autophagosome but the involvement of this protein in the fusion of the autophagosome and vacuole in plants remains unclear. It is suggested that the homologous yeast proteins Ykt6, Vam3, Ypt7, and complex HOPS are involved in the fusion of autophagosome and vacuole in plants. Additionally, it is suggested that plant proteins CHMP1, FREE1, VPS2.1, CFS1, and the complex EXO70B1 are involved in the autophagosome trafficking, autophagosome-vacuole fusion, and the release of the autophagic body into the vacuole lumen. The newly formed autophagic body inside the vacuole is rapidly degraded by lytic enzymes. One of them can be the vacuolar processing enzyme γ (VPEγ). Proteins involved in metabolite efflux from the vacuole to the cytoplasm during autophagy in plants have not been described so far. Only permease AVT3 was confirmed in Arabidopsis thaliana, but the involvement of this permease in the transport of metabolites coming from the degradation of the autophagic body is not confirmed. Question marks indicate the hypothetical involvement of plant proteins and complexes, or plant homologs of yeast proteins, during autophagy [[15]15].
Table 1. Proteins involved, or hypothetically involved, in trafficking and fusion of the autophagosome to the vacuole and formation of the autophagic body during macroautophagy in plants [[15]15] modified.
|
Protein |
Function |
References |
|
PI3P, Atg8 |
autophagosome trafficking and fusion |
[[20]] |
|
HOPS |
probably autophagosome-vacuole fusion |
|
|
VTI12 |
probably autophagosome formation, docking, and autophagosome-vacuole fusion, storage protein transport from cytoplasm to vacuole |
|
|
RABG3B |
autophagy enhancement during xylem development and pathogen-induced cell death, probably autophagosome formation and autophagosome-vacuole fusion |
|
|
CHMP1, FREE1, VPS2.1, CFS1, EXO70B1 |
probably autophagic trafficking, autophagosome-vacuole fusion, release of autophagic body |
[[8]] |
|
Protein |
Function |
References |
|
PI3P, Atg8 |
autophagosome trafficking and fusion |
[20] |
|
HOPS |
probably autophagosome-vacuole fusion |
[20,34] |
|
VTI12 |
probably autophagosome formation, docking, and autophagosome-vacuole fusion, storage protein transport from cytoplasm to vacuole |
[28,29,30,31] |
|
RABG3B |
autophagy enhancement during xylem development and pathogen-induced cell death, probably autophagosome formation and autophagosome-vacuole fusion |
[35,36] |
|
CHMP1, FREE1, VPS2.1, CFS1, EXO70B1 |
probably autophagic trafficking, autophagosome-vacuole fusion, release of autophagic body |
[8] |
REFERENCES
- Floyd, B.E.; Morriss, S.C.; MacIntosh, G.C.; Bassham, D.C. What to eat: Evidence for selective autophagy in plants. J. Integr. Plant Biol. 2012, 54, 907–920.
- Lv, X.; Pu, X.; Qin, G.; Zhu, T.; Lin, H. The roles of autophagy in development and stress responses in Arabidopsis thaliana. Apoptosis 2014, 19, 905–921.
- Rudnicka, K.W.; Szczęsna, E.; Miszczyk, E.; Mikołajczyk-Chmiela, M. Apoptosis and autophagy—Mechanisms and detection methods. Adv. Cell Biol. 2011, 38, 247–265.
- Hanamata, S.; Kurusu, T.; Kuchitsu, K. Roles of autophagy in male reproductive development in plants. Front. Plant Sci. 2014, 5, 457.
- Khan, M.S.; Hemalatha, S. Autophagy: Molecular insight and role in plant programmed cell death and defense mechanism. Int. Res. J. Biol. Sci. 2015, 4, 78–83.
- Fan, J.; Yu, L.; Xu, C. Dual role for autophagy in lipid metabolism in Arabidopsis. Plant Cell 2019, 31, 1598–1613.
- Reggiori, F.; Klionsky, D.J. Autophagic processes in yeast: Mechanism, machinery and regulation. Genetics 2013, 194, 341–361.
- Marshall, R.S.; Vierstra, R.D. Autophagy: The master of bulk and selective recycling. Annu. Rev. Plant Biol. 2018, 69, 173–208.
- Bozhkov, P.V. Plant autophagy: Mechanisms and functions. J. Exp. Bot. 2018, 69, 1281–1285.
- Ding, X.X.; Zhang, M.; Otegui, S. Plant autophagy: New flavors on the menu. Curr. Opin. Plant Biol. 2018, 46, 113–121.
- Puri, C.; Vicinanza, M.; Rubinsztein, D.C. Phagophores evolve from recycling endosomes. Autophagy 2018, 14, 1475–1477.
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222.
- Liu, Y.; Bassham, D.C. Autophagy: Pathways for Self-Eating in Plant Cells. Annu. Rev. Plant Biol. 2012, 63, 215–237.
- Masclaux-Daubresse, C.; Chen, Q.; Havé, M. Regulation of nutrient recycling via autophagy. Curr Opin Plant Biol. 2017, 39, 8–17.
- Stefaniak, S.; Wojtyla, Ł.; Pietrowska-Borek, M.; Borek, S. Completing autophagy: Formation and degradation of the autophagic body and metabolite salvage in plants. Int. J. Mol. Sci. 2020, 21, 2205.
- Monastyrska, I.; Rieter, E.; Klionsky, D.J.; Reggiori, F. Multiple roles of the cytoskeleton in autophagy. Biol. Rev. Camb. Philos. Soc. 2009, 84, 431–448.
- Winter, V.; Hauser, M.T. Exploring the ESCRTing machinery in eukaryotes. Trends Plant Sci. 2006, 11, 115–123.
- Gao, C.; Zhuang, X.; Cui, Y.; Fu, X.; He, Y.; Zhao, Q.; Jiang, L. Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc. Natl. Acad. Sci. USA 2015, 112, 1886–1891.
- Bassham, D.C. Plant autophagy—More than a starvation response. Curr. Opin. Plant Biol. 2007, 10, 587–593.
- Li, F.; Vierstra, R.D. Autophagy: A multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012, 17, 526–537.
- McEwan, D.G.; Popovic, D.; Gubas, A.; Terawaki, S.; Suzuki, H.; Stadel, D.; Dikic, I. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell 2015, 57, 39–54.
- Pankiv, S.; Alemu, E.A.; Brech, A.; Bruun, J.A.; Lamark, T.; Øvervatn, A.; Johansen, T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end–directed vesicle transport. J. Cell Biol. 2010, 188, 253–269.
- Jager, S. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 2004, 117, 4837–4848.
- Gutierrez, M.G.; Munafo, D.B.; Beron, W.; Colombo, M.I. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell Sci. 2004, 117, 2687–2697.
- Pankiv, S.; Johansen, T. FYCO1: Linking autophagosomes to microtubule plus end-directing molecular motors. Autophagy 2010, 6, 550–552.
- Lőrincz, P.; Kenéz, L.A.; Tóth, S.; Kiss, V.; Varga, Á.; Csizmadia, T.; Simon-Vecsei, Z.; Juhász, G. Vps8 overexpression inhibits HOPS-dependent trafficking routes by outcompeting Vps41/Lt. Elife 2019, 8, e45631.
- Nordmann, M.; Ungermann, C.; Cabrera, M. Role of rab7/ypt7 in organizing membrane trafficking at the late endosome. Rab GTPases and Membrane Trafficking. Curr. Biol. 2018, 28, 132–143.
- Surpin, M. The VTI Family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 2003, 15, 2885–2899.
- Surpin, M.; Raikhel, N. Traffic jams affect plant development and signal transduction. Nat. Rev. Mol. Cell Biol. 2004, 5, 100–109.
- Sanmartin, M.; Ordonez, A.; Sohn, E.J.; Robert, S.; Sanchez-Serrano, J.J.; Surpin, M.A.; Rojo, E. Divergent functions of VTI12 and VTI11 in trafficking to storage and lytic vacuoles in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 3645–3650.
- Bourdais, G.; McLachlan, D.H.; Rickett, L.M.; Zhou, J.; Siwoszek, A.; Häweker, H.; Robatzek, S. The use of quantitative imaging to investigate regulators of membrane trafficking in Arabidopsis stomatal closure. Traffic 2019, 20, 168–180.
- Kwon, S.I.; Cho, H.J.; Jung, J.H.; Yoshimoto, K.; Shirasu, K.; Park, O.K. The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J. 2010, 64, 151–164.
- Kwon, S.I.; Cho, H.J.; Kim, S.R.; Park, O.K. The Rab GTPase RabG3b Positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Physiol. 2013, 161, 1722–1736.
- Zientara-Rytter, K.; Sirko, A. To deliver or to degrade—An interplay of the ubiquitin-proteasome system, autophagy and vesicular transport in plants. FEBS J. 2016, 283, 3534–3555.
- Zhang, Z.; Feechan, A.; Pedersen, C.; Newman, M.A.; Qiu, J.; Olesen, K.L.; Thordal-Christensen, H. A SNARE-protein has opposing functions in penetration resistance and defence signalling pathways. Plant J. 2007, 49, 302–312.
- Uemura, T.; Ueda, T. Plant vacuolar trafficking driven by RAB and SNARE proteins. Curr. Opin. Plant Biol. 2014, 22, 116–121.