Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Catherine Yang and Version 1 by Zaibo Li.
HER2 intratumoral heterogeneity (ITH) is a well-known phenomenon in breast cancer, defined as the coexistence of subpopulations of tumor cells with different HER2 gene or protein expression within a tumor. HER2 ITH has been reported in up to 40% of breast cancers and to be associated with poor prognosis in patients with anti-HER2 targeted therapies and was proposed to be a potential mechanism for anti-HER2 resistance. HER2 ITH can be divided into non-genetic and genetic ITH based on different HER2 genetic amplification and genetic ITH has clustered, mosaic and scattered distribution patterns.
- HER2
- breast cancer
- heterogeneity
- immunohistochemistry
1. Introduction
Breast cancer is the most commonly diagnosed cancer and the leading cause of death in women worldwide. Globally, the estimated new breast cancer cases reached 2.26 million and accounted for 0.68 million cancer-related deaths in 2020 [1]. In 2023, newly diagnosed female breast cancers and the number of female breast cancer deaths were estimated to be 297,790 and 43,170 in the United States, respectively [2].
The ERBB2 (erb-b2 receptor tyrosine kinase 2) gene is a proto-oncogene encoding a tyrosine kinase receptor, human epidermal growth factor receptor-2 (HER2). HER2 is an orphan receptor with a constitutively activated conformation. It does not possess specific ligand or ligand binding activity. The HER2 protein can form homodimers when HER2 level is high or heterodimers with HER1, HER3 and HER4, but favors HER3. Dimerization of HER2 leads to phosphorylation of its tyrosine kinase domain and activates downstream oncogenic signaling pathways through PI3K/AKT and RAS/RAF/MEK/MAPK/MYC/c-jun to increase cell cycle progression, cell differentiation, survival, angiogenesis, tumorigenesis, migration and invasion [3,4,5][3][4][5]. HER2 overexpression or amplification is identified in approximately 15–20% of breast cancers [6,7][6][7] and is associated with higher histologic grade and stage, increased metastatic potential, decreased overall survival (OS), resistance to endocrine therapy and poor response to selected chemotherapy [8,9][8][9]. However, the introduction of anti-HER2 targeted therapies has profoundly changed the clinical course of HER2-positive cancer in both early and advanced disease, especially for metastatic breast cancer.
2. HER2 Assessment in Breast Cancer
Currently, the main application of HER2 assessment is in prediction of anti-HER2 treatment response in neoadjuvant and adjuvant settings. As a prognostic and predictive biomarker, HER2 status is routinely assessed by immunohistochemistry (IHC) and/or in situ hybridization (ISH) in breast cancer. In order to improve the accuracy of HER2 testing in invasive breast cancer, the American Society of Clinical Oncology and the College of American Pathologists (ASCO/CAP) first drafted a guideline for HER2 testing in breast cancer in 2007 [10]. The guideline recommended that HER2 status should be tested in all newly diagnosed invasive breast carcinoma. Originally, a HER2 positive result was defined as greater than 30% invasive tumor cells with uniform and intense circumferential staining by IHC or HER2 to centromeric enumeration probe for chromosome 17 (CEP17) ratio > 2.2 or average HER2 gene copy number greater than six per nucleus by ISH. In order to reduce the small number of potential false-negative results, ASCO/CAP revised the IHC criteria to more than 10% of invasive tumor cells in 2013 [11]. The ISH criteria for HER2 positive were also revised as HER2/CEP ratio ≥ 2.0 or HER2/CEP ratio < 2.0 with ≥6.0 HER2 copy number signals per nucleus. Testing was recommended for primary, recurrent and metastatic tumors. The ASCO/CAP further refined the guideline in 2018 [12]. Currently based on the 2018 guideline, breast cancers are classified as either HER2-positive (IHC3+ or 2+ with gene amplification by ISH) or HER2-negative (IHC 0+ or 1+ or 2+ without ISH amplification).
Several clinical trials, including the National Surgical Adjuvant Breast and Bowel Project (NSABP) B-31, the North Central Canter Treatment Group (NCCTG) N9831 and DESTINY-Breast04, demonstrated that a subset of breast cancer patients with HER2 low levels of expression also benefited from anti-HER2 targeted therapy, especially trastuzumab-deruxtecan (T-DXd) [13]. A new concept of “HER2 low” (IHC 1+ or 2+ without ISH amplification) breast cancer has been proposed [14,15,16][14][15][16]. Additional studies in this field may require a shift from the binary HER2 scoring system to a new system including HER2-positive, low and zero (IHC 0). Furthermore, breast cancer with HER2 ultra-low (IHC score 0 with incomplete and faint staining in ≤10% of tumor cells) has been highlighted [17]. The clinical trial investigating the potential benefit of T-DXd in HER2 ultra-low, hormone receptor positive breast cancer in metastatic setting is currently ongoing in DESTINY-Breast06. Currently, the assessment of HER2 low levels of expression in breast cancer has not been formally defined by ASCO/CAP.
3. HER2 Intratumoral Heterogeneity
Intratumoral heterogeneity (ITH) is defined as the coexistence of subpopulations of tumor cells that differ genetically, phenotypically or behaviorally within a primary tumor or between a primary tumor and its metastases. ITH poses a remarkable challenge for characterization of biomarkers and treatment selection.
HER2 ITH is a well-known phenomenon in breast cancer. The prevalence of HER2 heterogeneity has been reported in up to 40% of breast cancers [18,19,20,21,22,23,24,25,26,27][18][19][20][21][22][23][24][25][26][27]. It is rare in HER2 3+ cases, but significantly more common in HER2 equivocal cases [20,21,25,28,29,30][20][21][25][28][29][30]. Several studies also revealed remarkable HER2 ITH in HER2 low status [31].
HER2 ITH is defined by the co-existence of at least two distinct clones of cells with varying HER2 statuses within the same tumor. This means the different areas within the same tumor may have different levels of protein expression or gene amplification. HER2 ITH may present in three distinct patterns based on the geographic (spatial) distribution of heterogeneity: (1) clustered (regional) type, defined as two distinct areas with different HER2 gene amplified tumor cell populations; (2) mosaic (intermixed) type, defined as diffuse intermingling of cells with different HER2 gene amplification status; (3) scattered type, defined as isolated HER2 amplified tumor cells in a predominantly non-amplified tumor [19,32][19][32]. The clustered type is reported to be much less common compared to the mosaic type, 0.01% vs. 3% in unselected cohort and 4% vs. 15% of all IHC score 3+ and score 2+ cases subjected to ISH [33]. A genomic study with gene copy number profiling and massively parallel sequencing was investigated in 12 cases with HER2 ITH. It identified potential driver genetic alterations restricted to the HER2-negative cells, suggesting that HER2-negative components are likely driven by genetic alterations not present in the HER2-positive components, including BRF2 and DSN1 amplification and HER2 somatic mutations [34]. The mosaic and scattered types are more frequent and typically encountered in the HER2 2+ and ISH equivocal cases (HER2-double equivocal by 2013 guideline) [33]. Evaluation of HER2 status in these cases could be quite challenging. HER2 gene protein assay (GPA), a new HER2 testing modality with combined HER2 IHC and bright field ISH, enables assessment of HER2 protein expression and gene amplification simultaneously on a single slide and is particularly helpful in cases with HER2 ITH [27,35][27][35].
Tumor with genetic ITH shows co-existence of classic HER2-positive tumor cells with both HER2 gene amplification and HER2 protein overexpression, and HER2-negative tumor cells without HER2 gene amplification or HER2 protein overexpression. However, tumor with non-genetic ITH harbors tumor cells with HER2 gene amplification but no HER2 protein overexpression, and classic HER2-positive tumor cells (Figure 1). Tumor cells with such discordant HER2 gene amplification and HER2 protein expression can only be identified by using GPA, which shows IHC and ISH signals in the same tumor cells on one single slide. The different HER2 ITH patterns are summarized in Table 1.
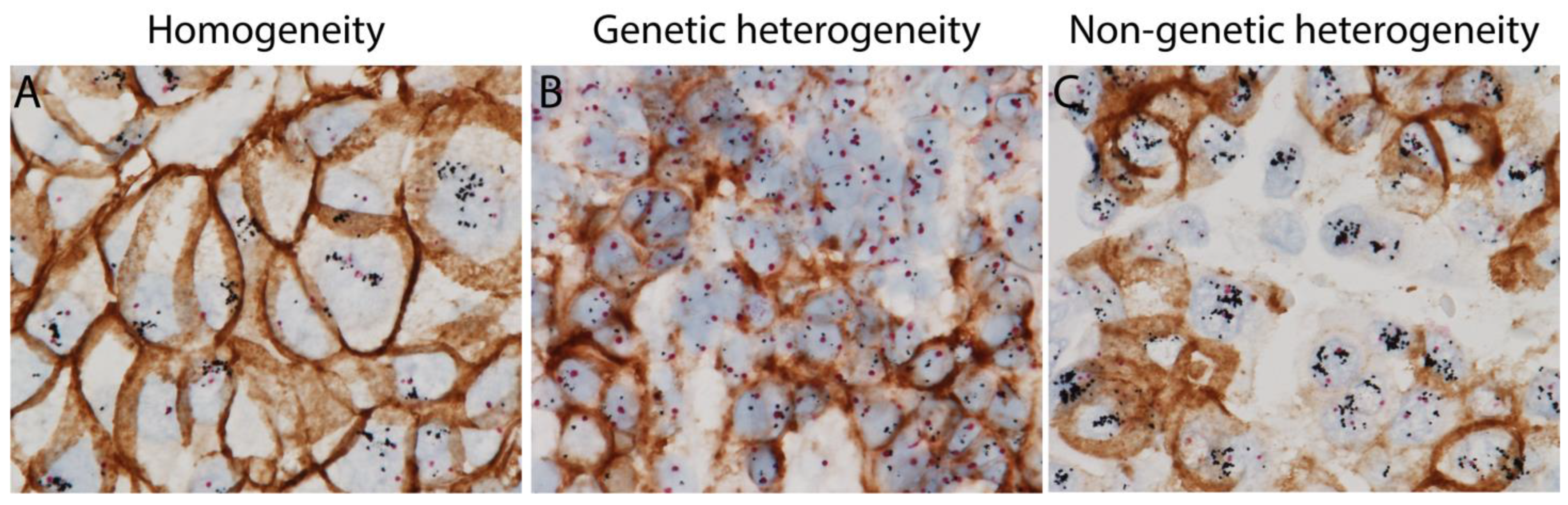
Figure 1. HER2 homogenous and heterogeneous staining patterns (intratumoral heterogeneity (ITH)) using HER2 gene protein assay (GPA) with simultaneous HER2 in situ hybridization and immunohistochemistry on breast cancer tissue sections. (A) Homogeneous staining with classic HER2 positive tumor cells with both amplified HER2 gene and overexpressed HER2 protein. (B) HER2 genetic intratumoral heterogeneity with classic HER2 positive tumor cells and HER2 negative tumor cells. (C) HER2 non-genetic intratumoral heterogeneity with a mixture of classic HER2 positive tumor cells and non-classic HER2 positive tumor cells harboring amplified HER2 gene, but no overexpression of HER2 protein.
Table 1.
Summary of HER2 intratumoral heterogeneity patterns.
| HER2 ITH Patterns | Definition | |
|---|---|---|
| Genetic | Clustered Type | Two distinct areas with different HER2 gene amplification |
| Mosaic Type | Diffuse intermingling of cells with different HER2 amplification status | |
| Scattered Type | Isolated HER2 amplified tumor cells in a predominantly non-amplified tumor | |
| Non-genetic | Tumor cells with HER2 gene amplification without HER2 protein expression intermixed with tumor cells with concordant HER2 amplification and protein expression | |
The difference of HER2 expression levels between primary and residual tumors after neoadjuvant therapies has been reported. The potential mechanism could be that the HER2-positive cells are more sensitive to anti-HER2 targeted therapies which lead to selected proliferation of clones with HER2-negative or low expression [36]. The discordance of HER2 status has also been reported in 0–34% of breast cancers between the primary and metastatic sites [37]. Possible explanations for this wide range of variation could be due to differences in fixation and ischemic time, different patient populations included in study cohorts, different methodologies, and different HER2 antibody clones in different studies. It could also be due to inter-observer interpretation variabilities, especially in tumors with HER2 ITH, as discussed later. The potential mechanisms of loss of HER2 expression in metastases include genetic drift, clonal selection during tumor progression, ITH, aforementioned anti-HER2 therapeutic selection or possible discrepant biomarker testing results [37]. Discordance of HER2 expression between primary and either residual tumor or metastatic settings was associated with poor prognosis or a lack of pathologic complete response (pCR) [37,38][37][38].
5. Clinicopathologic Features of Breast Cancer with HER2 ITH
HER2 ITH exists in HER2-low breast cancers more frequently than HER2-zero or HER2 positive cancers, featuring a pattern of diffuse intermingling of HER2-positive and HER2-negative tumor cells (mosaic pattern) [31,41,42][31][39][40]. The majority of these cases are hormone receptor positive [43][41]. In contrast to HER2-positive cases, HER2 ITH cases are associated with lower histologic grade, smaller tumor size with a biologic features resembling HER2-negative cases [30]. Among the HER2-negative carcinomas, cases with ITH were associated with larger size, higher grade and greater incidence of lymph node metastasis [29].References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249.
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48.
- Baselga, J. Why the epidermal growth factor receptor? The rationale for cancer therapy. Oncologist 2002, 7 (Suppl. S4), 2–8.
- Puglisi, F.; Fontanella, C.; Amoroso, V.; Bianchi, G.V.; Bisagni, G.; Falci, C.; Fontana, A.; Generali, D.; Gianni, L.; Grassadonia, A.; et al. Current challenges in HER2-positive breast cancer. Crit. Rev. Oncol. Hematol. 2016, 98, 211–221.
- Hayes, D.F. HER2 and Breast Cancer—A Phenomenal Success Story. N. Engl. J. Med. 2019, 381, 1284–1286.
- Cronin, K.A.; Harlan, L.C.; Dodd, K.W.; Abrams, J.S.; Ballard-Barbash, R. Population-based estimate of the prevalence of HER-2 positive breast cancer tumors for early stage patients in the US. Cancer Investig. 2010, 28, 963–968.
- Guarneri, V.; Barbieri, E.; Dieci, M.V.; Piacentini, F.; Conte, P. Anti-HER2 neoadjuvant and adjuvant therapies in HER2 positive breast cancer. Cancer Treat. Rev. 2010, 36 (Suppl. S3), S62–S66.
- Menard, S.; Tagliabue, E.; Campiglio, M.; Pupa, S.M. Role of HER2 gene overexpression in breast carcinoma. J. Cell. Physiol. 2000, 182, 150–162.
- Abd El-Rehim, D.M.; Pinder, S.E.; Paish, C.E.; Bell, J.A.; Rampaul, R.S.; Blamey, R.W.; Robertson, J.F.; Nicholson, R.I.; Ellis, I.O. Expression and co-expression of the members of the epidermal growth factor receptor (EGFR) family in invasive breast carcinoma. Br. J. Cancer 2004, 91, 1532–1542.
- Wolff, A.C.; Hammond, M.E.; Schwartz, J.N.; Hagerty, K.L.; Allred, D.C.; Cote, R.J.; Dowsett, M.; Fitzgibbons, P.L.; Hanna, W.M.; Langer, A.; et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J. Clin. Oncol. 2007, 25, 118–145.
- Wolff, A.C.; Hammond, M.E.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.M.; Bilous, M.; Fitzgibbons, P.; et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. 2013, 31, 3997–4013.
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.S.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W.; et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2018, 36, 2105–2122.
- MacNeil, I.A.; Burns, D.J.; Rich, B.E.; Soltani, S.M.; Kharbush, S.; Osterhaus, N.G.; Sullivan, B.F.; Hawkins, D.M.; Pietruska, J.R.; Laing, L.G. New HER2-negative breast cancer subtype responsive to anti-HER2 therapy identified. J. Cancer Res. Clin. Oncol. 2020, 146, 605–619.
- Modi, S.; Jacot, W.; Yamashita, T.; Sohn, J.; Vidal, M.; Tokunaga, E.; Tsurutani, J.; Ueno, N.T.; Prat, A.; Chae, Y.S.; et al. Trastuzumab Deruxtecan in Previously Treated HER2-Low Advanced Breast Cancer. N. Engl. J. Med. 2022, 387, 9–20.
- Zhang, H.; Katerji, H.; Turner, B.M.; Hicks, D.G. HER2-Low Breast Cancers. Am. J. Clin. Pathol. 2022, 157, 328–336.
- Banerji, U.; van Herpen, C.M.L.; Saura, C.; Thistlethwaite, F.; Lord, S.; Moreno, V.; Macpherson, I.R.; Boni, V.; Rolfo, C.; de Vries, E.G.E.; et al. Trastuzumab duocarmazine in locally advanced and metastatic solid tumours and HER2-expressing breast cancer: A phase 1 dose-escalation and dose-expansion study. Lancet Oncol. 2019, 20, 1124–1135.
- Venetis, K.; Crimini, E.; Sajjadi, E.; Corti, C.; Guerini-Rocco, E.; Viale, G.; Curigliano, G.; Criscitiello, C.; Fusco, N. HER2 Low, Ultra-low, and Novel Complementary Biomarkers: Expanding the Spectrum of HER2 Positivity in Breast Cancer. Front. Mol. Biosci. 2022, 9, 834651.
- Vance, G.H.; Barry, T.S.; Bloom, K.J.; Fitzgibbons, P.L.; Hicks, D.G.; Jenkins, R.B.; Persons, D.L.; Tubbs, R.R.; Hammond, M.E.; College of American P. Genetic heterogeneity in HER2 testing in breast cancer: Panel summary and guidelines. Arch Pathol. Lab. Med. 2009, 133, 611–612.
- Hanna, W.M.; Ruschoff, J.; Bilous, M.; Coudry, R.A.; Dowsett, M.; Osamura, R.Y.; Penault-Llorca, F.; van de Vijver, M.; Viale, G. HER2 in situ hybridization in breast cancer: Clinical implications of polysomy 17 and genetic heterogeneity. Mod. Pathol. 2014, 27, 4–18.
- Lee, H.J.; Seo, A.N.; Kim, E.J.; Jang, M.H.; Suh, K.J.; Ryu, H.S.; Kim, Y.J.; Kim, J.H.; Im, S.A.; Gong, G.; et al. HER2 heterogeneity affects trastuzumab responses and survival in patients with HER2-positive metastatic breast cancer. Am. J. Clin. Pathol. 2014, 142, 755–766.
- Allison, K.H.; Dintzis, S.M.; Schmidt, R.A. Frequency of HER2 heterogeneity by fluorescence in situ hybridization according to CAP expert panel recommendations: Time for a new look at how to report heterogeneity. Am. J. Clin. Pathol. 2011, 136, 864–871.
- Chang, M.C.; Malowany, J.I.; Mazurkiewicz, J.; Wood, M. ‘Genetic heterogeneity’ in HER2/neu testing by fluorescence in situ hybridization: A study of 2,522 cases. Mod. Pathol. 2012, 25, 683–688.
- Hanna, W.; Nofech-Mozes, S.; Kahn, H.J. Intratumoral heterogeneity of HER2/neu in breast cancer—A rare event. Breast J. 2007, 13, 122–129.
- Bartlett, A.I.; Starcyznski, J.; Robson, T.; Maclellan, A.; Campbell, F.M.; van de Velde, C.J.; Hasenburg, A.; Markopoulos, C.; Seynaeve, C.; Rea, D.; et al. Heterogeneous HER2 gene amplification: Impact on patient outcome and a clinically relevant definition. Am. J. Clin. Pathol. 2011, 136, 266–274.
- Seol, H.; Lee, H.J.; Choi, Y.; Lee, H.E.; Kim, Y.J.; Kim, J.H.; Kang, E.; Kim, S.W.; Park, S.Y. Intratumoral heterogeneity of HER2 gene amplification in breast cancer: Its clinicopathological significance. Mod. Pathol. 2012, 25, 938–948.
- Hou, Y.; Nitta, H.; Wei, L.; Banks, P.M.; Portier, B.; Parwani, A.V.; Li, Z. HER2 intratumoral heterogeneity is independently associated with incomplete response to anti-HER2 neoadjuvant chemotherapy in HER2-positive breast carcinoma. Breast Cancer Res. Treat. 2017, 166, 447–457.
- Hou, Y.; Nitta, H.; Li, Z. HER2 Gene Protein Assay Is Useful to Determine HER2 Status and Evaluate HER2 Heterogeneity in HER2 Equivocal Breast Cancer. Am. J. Clin. Pathol. 2017, 147, 89–95.
- Ohlschlegel, C.; Zahel, K.; Kradolfer, D.; Hell, M.; Jochum, W. HER2 genetic heterogeneity in breast carcinoma. J. Clin. Pathol. 2011, 64, 1112–1116.
- Shafi, H.; Astvatsaturyan, K.; Chung, F.; Mirocha, J.; Schmidt, M.; Bose, S. Clinicopathological significance of HER2/neu genetic heterogeneity in HER2/neu non-amplified invasive breast carcinomas and its concurrent axillary metastasis. J. Clin. Pathol. 2013, 66, 649–654.
- Yang, Y.L.; Fan, Y.; Lang, R.G.; Gu, F.; Ren, M.J.; Zhang, X.M.; Yin, D.; Fu, L. Genetic heterogeneity of HER2 in breast cancer: Impact on HER2 testing and its clinicopathologic significance. Breast Cancer Res. Treat. 2012, 134, 1095–1102.
- Miglietta, F.; Griguolo, G.; Bottosso, M.; Giarratano, T.; Lo Mele, M.; Fassan, M.; Cacciatore, M.; Genovesi, E.; De Bartolo, D.; Vernaci, G.; et al. Evolution of HER2-low expression from primary to recurrent breast cancer. NPJ Breast Cancer 2021, 7, 137.
- Marchio, C.; Annaratone, L.; Marques, A.; Casorzo, L.; Berrino, E.; Sapino, A. Evolving concepts in HER2 evaluation in breast cancer: Heterogeneity, HER2-low carcinomas and beyond. Semin. Cancer Biol. 2021, 72, 123–135.
- Grassini, D.; Cascardi, E.; Sarotto, I.; Annaratone, L.; Sapino, A.; Berrino, E.; Marchio, C. Unusual Patterns of HER2 Expression in Breast Cancer: Insights and Perspectives. Pathobiology 2022, 89, 278–296.
- Ng, C.K.; Martelotto, L.G.; Gauthier, A.; Wen, H.C.; Piscuoglio, S.; Lim, R.S.; Cowell, C.F.; Wilkerson, P.M.; Wai, P.; Rodrigues, D.N.; et al. Intra-tumor genetic heterogeneity and alternative driver genetic alterations in breast cancers with heterogeneous HER2 gene amplification. Genome Biol. 2015, 16, 107.
- Nitta, H.; Kelly, B.D.; Allred, C.; Jewell, S.; Banks, P.; Dennis, E.; Grogan, T.M. The assessment of HER2 status in breast cancer: The past, the present, and the future. Pathol. Int. 2016, 66, 313–324.
- Morganti, S.; Ivanova, M.; Ferraro, E.; Ascione, L.; Vivanet, G.; Bonizzi, G.; Curigliano, G.; Fusco, N.; Criscitiello, C. Loss of HER2 in breast cancer: Biological mechanisms and technical pitfalls. Cancer Drug Resist. 2022, 5, 971–980.
- Hou, Y.; Shen, R.; Chaudhary, S.; Gao, F.; Li, Z. Correlation of Expression of Breast Biomarkers in Primary and Metastatic Breast Carcinomas: A Single-Institution Experience. Acta Cytol. 2016, 60, 481–489.
- Caswell-Jin, J.L.; McNamara, K.; Reiter, J.G.; Sun, R.; Hu, Z.; Ma, Z.; Ding, J.; Suarez, C.J.; Tilk, S.; Raghavendra, A.; et al. Clonal replacement and heterogeneity in breast tumors treated with neoadjuvant HER2-targeted therapy. Nat. Commun. 2019, 10, 657.
- Sapino, A.; Goia, M.; Recupero, D.; Marchio, C. Current Challenges for HER2 Testing in Diagnostic Pathology: State of the Art and Controversial Issues. Front. Oncol. 2013, 3, 129.
- Denkert, C.; Seither, F.; Schneeweiss, A.; Link, T.; Blohmer, J.U.; Just, M.; Wimberger, P.; Forberger, A.; Tesch, H.; Jackisch, C.; et al. Clinical and molecular characteristics of HER2-low-positive breast cancer: Pooled analysis of individual patient data from four prospective, neoadjuvant clinical trials. Lancet Oncol. 2021, 22, 1151–1161.
- Filho, O.M.; Viale, G.; Stein, S.; Trippa, L.; Yardley, D.A.; Mayer, I.A.; Abramson, V.G.; Arteaga, C.L.; Spring, L.M.; Waks, A.G.; et al. Impact of HER2 Heterogeneity on Treatment Response of Early-Stage HER2-Positive Breast Cancer: Phase II Neoadjuvant Clinical Trial of T-DM1 Combined with Pertuzumab. Cancer Discov. 2021, 11, 2474–2487.
More
