Proteolytic enzymes, also known as peptidases, are one of the most abundant groups of enzymes in living organisms. They control the activation, synthesis and turnover of proteins, and regulate most biochemical and physiological processes, such as digestion, fertilization, growth, differentiation, cell signaling/migration, immunological defense, wound healing, and apoptosis. They are consequently major regulators of homeostasis, ageing, and different human diseases like cancer, hypertension, diabetes, inflammation, neurodegeneration, Alzheimer among others. Proteases are also essential for propagation of infectious agents, being major contributors of pathogenesis in several infectious diseases, including the current coronavirus emergent pandemic SARS COVID 19. Among peptidases, aminopeptidases catalyze the cleavage of the N-terminal amino acids of proteins or peptide substrates. They are distributed in many phyla and play critical roles in physiology and pathophysiology. Many of them are metallopeptidases belonging to the M1 and M17 families, among others. Some, such as M1 aminopeptidases N and A, thyrotropin-releasing hormone-degrading ectoenzyme, and M17 leucyl aminopeptidase, are targets for the development of therapeutic agents for human diseases, including cancer, hypertension, central nervous system disorders, inflammation, immune system disorders, skin pathologies, and infectious diseases, such as malaria.
1. Metallopeptidases: General Characteristics and Classification
Metallopeptidases constitute the most diverse catalytic type within proteases, since they include both endopeptidases and exopeptidases, cytosolic enzymes, and others that are secreted to the outside of cells, as well as enzymes associated with the plasma membrane and cell organelles. They are widely distributed in all forms of life such as viruses, bacteria, fungi, and plant and animal cells, indicating the important role they play in biological processes.
Metalloproteases are included among the hydrolases in which the nucleophilic attack on the peptide bond is mediated by a water molecule. This is a feature they share with aspartic-type peptidases, but in metallopeptidases, a divalent metal cation activates the water molecule
[1][63]. This divalent cation is usually zinc (Zn
2+) but can sometimes be cobalt (Co
2+) or manganese (Mn
2+). The metal ion is held in the protein structure by amino acids that act as ligands.
Metallopeptidases can be divided into two large groups based on the number of metal ions required for catalysis. In many metallopeptidases, only one metal ion is required, which frequently is Zn2+; however, there is another group of families in which two cocatalytically acting metal ions are required. Within this group, there are families that have two Zn2+ ions and all the families in which Co2+ or Mn2+ are essential for catalysis. In families where only one metal ion acts, three amino acid residues are required to act as metal ligand coordinators, and in families with cocatalytic ions, only five amino acids are required since one of them acts as a ligand coordinator for both metal ions. All metallopeptidases with cocatalytic ions are exopeptidases, while metallopeptidases with a single metal ion can be both exo- and endopeptidases.
Various attempts have been made to classify proteases. The most accepted today is the one initially proposed by Rawlings and Barrett
[2][3][64,65] that is continuously updated at (
https://www.ebi.ac.uk/merops/, accessed on 9 January 2023)
[4][1]. From the general classification of the nine mechanistic classes, they first group the enzymes of each class into families. A family is defined as a group of (homologous) peptidases in which each member shows significant amino acid sequence identity with the “type enzyme” or at least with another member of the family homologous to the type enzyme, mainly in the region of the peptidase that is related to its catalytic activity. The selection criteria used by these authors were very strict, in such a way that they guarantee a common ancestor for the members of a family, which are, therefore, homologous according to the definition of Reeck et al.
[5][66]. Each family is named with a letter denoting the catalytic type (Example: M for metallopeptidases), followed by an arbitrarily assigned number. At a higher level of hierarchy, we find the clan, which is the term used by these authors to describe a group of families, whose members originate from a common ancestor protein but which have diverged to a point where relationships between them cannot be demonstrated by homology in their primary structures. The main evidence for the clan level is the relationship between families in terms of similarities in the three-dimensional structure of their members, in the arrangement of catalytic residues in the peptide structure, as well as similarities in the amino acid sequence around the catalytic residues
[4][1].
Up to now, 16 clans of metallopeptidases have been described: MA, MC, MD, ME, MF, MG, MH, MJ, MM, MN, MO, MP, MQ, MS, MT, and MU, with six of them comprising exo-peptidases. Overall, they form 76 families, with clans MA and MF being two of the most well characterized with enzymes from all living organisms
[4][1].
2. Clan MA: Subclan MA (E)
The clan MA is the largest of the metallopeptidases, with a total of 49 families
[4][1], all consisting of enzymes that contain a single Zn
2+ in their active sites. This clan is made up of both endopeptidases and exopeptidases, comprising aminopeptidases (families M1, M2, M4, M5, M9, M13, M30, M36, M48, and M61), carboxypeptidases (M2 and M32), peptidyl-dipeptidases (M2), oligopeptidases (M3 and M13), and endopeptidases (families M4, M10, and M12). In the enzymes of the MA clan, the Zn
2+ atom is coordinated to the protein through two His residues, which are part of the HEXXH motif. In addition to the His residues, the catalytic Zn
2+ is coordinated by a water molecule and a third residue, the nature of which determines the clan’s subdivision into the MA (E) and MA (M) subclans. In the subclan MA (M), the third ligand can be a residue of His or Asp within the HEXXHXXGXXH/D signature sequence, while in subclan MA (E) the third ligand is a residue of Glu, located at least 14 residues after the carboxyl terminus of the HEXXH motif
[4][1] (
Figure 1). The oxygen atom of the water molecule that acts as a metal ligand is the nucleophilic agent that attacks the carbonyl of the peptide bond to be hydrolyzed.
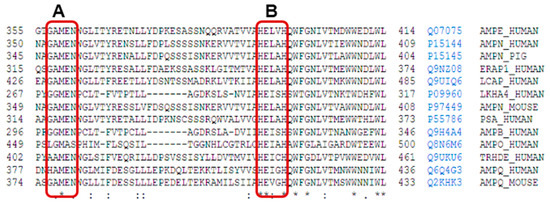
Figure 1. Uniprot alignment of the aminoacidic sequence of the active site of various M1 family aminopeptidases: AMPE_Human: human aminopeptidase E, AMPN_Human: human aminopeptidase N, AMPN_Pig: porcine aminopeptidase N, ERAP1_Human: endoplasmatic reticulum aminopeptidase 1, LCAP_Human: human leucyl-cystinyl aminopeptidase, LKHA4_Human: human leukotriene A4 hydrolase, PSA_Human: human puromycin sensitive aminopeptidase, AMPB_Human: human aminopeptidase B, AMPO_Human: human aminopeptidase O, TRHDE_Human: human thyrotrophin-releasing hormone-degrading enzyme or pyroglutamyl aminopeptidase II, AMPQ_Human: human aminopeptidase Q, AMPQ_Mouse: mouse aminopeptidase Q. On the right of each sequence, the access number and identifiers from Uniprot are included. The short name for each enzyme corresponds to Uniprot abbreviations. The rectangle A encircles the conserved sequence GAMEN related with the aminopeptidase activities of these enzymes from M1 family, and the rectangle B encircles the consensus sequences HEXXH from the active site. Signs below alignment points to other highly conserved amino acid residues inside the M1 family. * indicates residues completely conserved, : indicates position with high degree of conservation of the residues, and . indicates position with mild degree of conservation of the residues.
Thermolysin (EC 3.4.24.27), a secretory endopeptidase, is the model enzyme of the MA clan and its structure, widely characterized, is a point of reference for the study of the enzymes of this clan due to the high structural similarity between them in terms of the organization of the active center
[4][1]. Among the most studied families of the subclan MA (E) is M1, whose members show a wide distribution in the living world (
Table 1); furthermore, they are involved in many functions that include cell maintenance, growth, development, and defense
[6][8]. This family includes enzymes of Gram (+) and Gram (−) bacteria, cyanobacteria, archaea, protozoa, fungi, animals, and plants
[4][6][1,8].
,75,76]; human APA
[19][77]; human ERAP-1
[20][78]; and porcine and human APN in complex with substrates and bestatin
[12][21][70,79], among others (
Table 2) (
Figure 2). In all these structures, it can be seen that the catalytic domain of this enzymatic family presents a high structural similarity with thermolysin, despite the fact that in some cases, there is only 7% identity in sequence with the corresponding polypeptide chains
[13][71]. The high availability of M1 aminopeptidase structures, the well-studied active site able to the binding of small molecules, and the well characterized reaction mechanisms, make M1 aminopeptidases ideal candidates for the application of structure-guided inhibitor discovery, including high-throughput screenings in different databases of marine and other natural compounds. These inhibitors have potentialities in different infectious and chronic human diseases
[22][23][80,81].
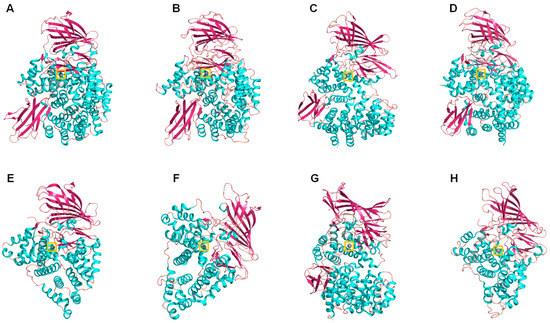
Figure 2. Cartoon representation of different M1 family aminopeptidases: (A) pepN from Escherichia coli (PDB ID: 2dq6), (B) Plasmodium falciparum aminopeptidase N PfA-M1 (PDB ID: 3ebh), (C) human ERAP 1 (PDB ID: 3mdj), (D) human ERAP 2 (PDB ID: 3se6), (E) human leukotriene A4 hydrolase (PDB ID: 1hs6), (F) Saccharomyces cerevisiae leukotriene A4 hydrolase (PDB ID: 2xq0), (G) Thermoplasma acidophilum tricorn interacting factor F3 (PDB ID: 1z1w), (H) Colwellia psychrerythraea cold-active aminopeptidase (PDB ID: 3cia). Colors: alpha-helices (cyan), beta sheets (warm pink), and loops (salmon). The zinc atoms are shown as gray spheres highlighted in a yellow box.
3. M1 Family of Mmetallo Aaminopeptidases
The aminopeptidases of the M1 family exist in monomeric or dimeric forms. In eukaryotes, they are generally membrane-associated enzymes such as mammalian APN (i.e., from human or pig), acidic or glutamyl aminopeptidase (APA), adipocyte-derived leucine aminopeptidase, and thyrotropin-releasing hormone-degrading ectoenzyme (TRH-DE), also known as pyroglutamyl peptidase II
[7][16]. Some are cytosolic enzymes, such as leukotriene A4 hydrolase (bifunctional enzyme with aminopeptidase activity)
[8][67] and aminopeptidase B (APB)
[9][68], or associated with the cell wall
[10][9], such as the neutral aminopeptidase (APN, EC 3.4.11.2) of the yeast
Candida albicans [11][69]. The structure of the membrane-bound aminopeptidases of the M1 family, in general, comprises a short intracellular tail attached to the transmembrane domain and a large ectodomain formed, in turn, by 2- or 3-folded and conserved domains. Domain I, N-terminal, has a β-sheet nucleus that, although it is widely exposed to the solvent, contains a hydrophobic region that continues in an anchorage region in the membrane. Catalytic domain II, such as that of thermolysin, contains an active site flanked by a mixed structure of β-sheet and α-helix that is highly conserved throughout the family. Domain III, which is composed of an immunoglobulin-like fold, does not appear in some family members (such as leukotriene A4 hydrolase). Domain IV, C-terminal, is the most variable region within the family. It is completely helical, with such an arrangement that it covers the active site; it is also involved in the dimerization of the mammalian isoforms
[6][8]. Disulfide bridges and abundant glycosylations are generally seen in this extracellular region, and some of these enzymes are surface antigens
[4][7][1,16].
In the M1 family, a well-conserved motif is the Gly-Ala/X-Met-Glu-Asn (GAMEN/GXMEN) sequence. This sequence, also known as the exopeptidase motif, frequently shows variations in the first two residues, and is very useful for the identification of family members
[6][7][12][8,16,70] (
Figure 1).
Through the technique of crystallography and X-ray diffraction, the three-dimensional structures of several members of this family have been elucidated, such as leukotriene A4 hydrolase in complex with its inhibitor bestatin
[13][71]; tricorn-interacting factor 3 of
Thermoplasma acidophilum [14][72];
Escherichia coli APN (Pep N) in complex with its inhibitor bestatin
[15][73];
Plasmodium falciparum (PfA-M1) alone and in complex with bestatin and low molecular mass analogs
[16][17][18][74