
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Francesca Di Nunzio | + 1461 word(s) | 1461 | 2021-02-24 04:59:08 | | | |
| 2 | Vicky Zhou | Meta information modification | 1461 | 2021-02-26 05:25:13 | | |
Video Upload Options
HIV-1 capsid has been recognized to have an important role as a structural protein that holds the viral genome, together with viral proteins essential for viral life cycle, such as the reverse transcriptase (RT) and the integrase (IN). The reverse transcription process takes place between the cytoplasm and the nucleus of the host cell, thus the Reverse Transcription Complexes (RTCs)/Pre-integration Complexes (PICs) are hosted in intact or partial cores. Early biochemical assays failed to identify the viral CA associated to the RTC/PIC, possibly due to the stringent detergent conditions used to fractionate the cells or to isolate the viral complexes. More recently, it has been observed that some host partners of capsid, such as Nup153 and CPSF6, can only bind multimeric CA proteins organized in hexamers. Those host factors are mainly located in the nuclear compartment, suggesting the entrance of the viral CA as multimeric structure inside the nucleus. Recent data show CA complexes within the nucleus having a different morphology from the cytoplasmic ones, clearly highlighting the remodeling of the viral cores during nuclear translocation. Thus, the multimeric CA complexes lead the viral genome into the host nuclear compartment, piloting the intranuclear journey of HIV-1 in order to successfully replicate.
1. Introduction
Human immunodeficiency virus 1 (HIV-1) is part of the lentiviruses subfamily that disseminated around humans starting from the twentieth century; nonetheless, the virus was isolated only in 1983 [1]. The main outcome of HIV-1 infection is the deep depletion of CD4+ T lymphocytes; however, the count decrease is just transient during the first weeks of infection, making complex the early diagnosis. The T count reduction slowly worsens over the years, yielding to the Acquired Immune Deficiency Syndrome (AIDS) and the related consequences.
The key feature of lentiviruses consists in the ability to reverse transcribe their RNA genome into double-stranded DNA with subsequent integration into the host chromatin [2][3]. Usually, HIV-1 integration step targets active host genes to ensure the release of its own progeny, but some not yet clear conditions favor the persistence of silent viral genomes (a process known as latency) [4]. Indeed, the virus survives silently in apparently healthy cells, making it difficult to cure AIDS. For their importance in the viral life cycle, the reverse transcriptase (RT) and the integrase (IN) have always been in the spotlight as crucial partners of the reverse-transcribed DNA and as therapeutic targets [5][6]. However, from the past years, the viral capsid progressively gained relevance both in reverse transcription and post-nuclear entry steps [7][8][9][10][11][12], but also as a target for new anti-retroviral treatments [13][14]. Indeed, the scientific community is abandoning the early and absolutistic view of the immediate uncoating, likely arisen from the difficulties in studying the association of viral capsid with the Reverse Transcription Complexes/Pre-integration Complexes (RTC/PIC) by biochemical essays [15][16].
Thanks to new cutting-edge technologies to study the fate of the capsid in infected cells, it has now been put forward the idea of a more tightly regulated uncoating process [11], in which the core shell is preserved until the nuclear translocation step [17]. Importantly, the progressive uncoating ensures protection of the viral complexes from the cytoplasmic environment, and it plays a key direct or indirect role in DNA synthesis, nuclear import, and integration. On one side, in vitro studies are essential for the characterization of the RTC and PIC, being that these transient and heterogeneous viral structures are very difficult to study in cells. On the other side, in vitro studies as biochemical approaches of purification of RTC/PIC [15][16][18] were unable to unravel the role of the capsid in HIV-1 life cycle. These results were mainly due to the cell-free experimental context or for the usage of strong detergents. In recent years, the advent of new technologies, particularly in the imaging field, allowed to develop a better overview of the role of the viral capsid, not only for its structural importance, but also as a key viral component for the RTC/PIC dynamics in the cells.
2. The Role of the HIV-1 CA in the Viral Integration
A role of the CA in HIV-1 integration has been envisaged as the depletion of the Nups capable of binding the viral CA hexamers results in a change in the distribution of integration sites [19]. However, it is still not clear whether the viral CA plays a direct role in the HIV-1 integration. Probably, nuclear uncoating could represent an advantage for HIV-1 nuclear steps. Viral CA complexes can protect the viral genome from antiviral sensors and escort the mature PIC to the vicinity of active gene regions for efficient viral integration. Following in live HIV-1 labelled CA in HeLa cells, it is possible to see the disappearance of CA signal at the nuclear location where an HIV-1 transcriptional focus appears [20]. Then, how does HIV-1 CA know where to locate the viral genetic material? CPSF6-CA interaction [21] seems to dictate the nuclear location of HIV-1 genomes [22]. Importantly, through the direct detection of HIV-1 DNA, it was possible to visually confirm the relevance of this interaction, which might have a role in HIV integration sites distribution [23][24][25][26]. New results demonstrate that EdU-labelled viral DNA [27], as well as viral IN [22][28], accumulate in CPSF6 clusters, which are retained in SC35-positive nuclear speckles [22][28]. Therefore, CPSF6 seems to be not only responsible for HIV-1 CA shuttling, but also to escort the RTC/PIC in nuclear speckles (NS) regions where the viral DNA labeled with EdU has been found [22][27]. Of note, the NS regions are known to be interchromatin granules [27][29]; therefore, they unlikely can be sites of viral integration. However, NSs have been indicated as HIV-1 integration sites because of the detection in these nuclear organelles components of the P-TEFb complex [22], which are usually recruited to the transcribing viral genome [30]. On the other hand, these factors were found by other authors in the vicinity of NSs [29]. NSs are also considered nuclear storage sites, rather than sites of functional processes [29][31]. However, what determines the specific subset of transcription factors localized to nuclear speckles is still unclear. Mainly, NSs are important for the assembly of higher-order complexes and/or for the state or accessibility of splicing and transcription factors [29]. Indeed, NS-neighboring chromatin regions that contain active chromatin [32] represent favorable chromatin loci for viral integration [28] (Figure 1).
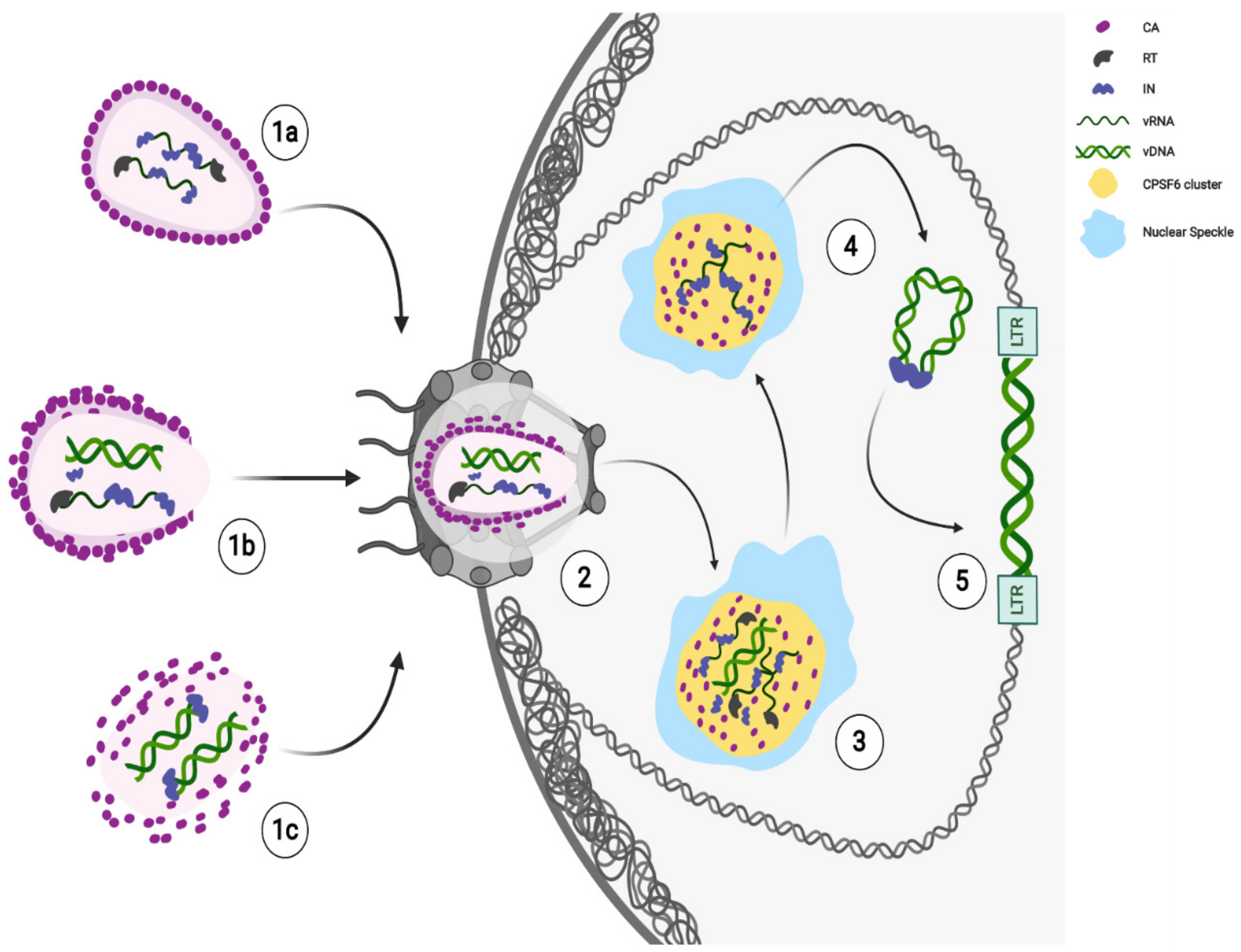
Recent findings show late HIV-1 reverse-transcribed DNA separated from IN foci [28][33] in the nucleus. The majority of intranuclear-detected INs are retained in CPSF6 clusters, while the integration of the viral DNA occurs in proximity, but not inside these nuclear condensates [28] (Figure 1). Thus, the HIV-induced CPSF6/SC35 membraneless organelles (HIV-1 MLOs) might be safe nuclear sites to complete reverse transcription and uncoating. Again, the accumulation of RTC in CPSF6/NS appears to precede the completion or, possibly, the beginning of DNA synthesis. Concentration of forming PICs in NS may promote integration of HIV-1 into euchromatin regions located outside, but not far from NS, for optimal replication [28].
3. Conclusions
The knowledge of HIV-1 capsid function deeply evolved from the mere structural-shield protein. Recent exciting results about nuclear uncoating and nuclear reverse transcription completely change our view on HIV-1 early phases of the life cycle. Apparently, nuclear import precedes the complete uncoating and the reverse transcription can occur in nuclear HIV-specific membraneless organelles (HIV-1 MLOs), at least in macrophages. Does the nuclear reverse transcription represent an advantage for HIV? Is it correlated to an efficient replication? All these questions open new frontiers of research based on the role of HIV-1 MLOs on viral persistence and rebound, which represent the major obstacle to cure AIDS.
Possibly, the nuclear CA can trigger nuclear antiviral pathways [34], but a late uncoating keeps safe the viral genetic material until integration. Future studies are required to clarify whether the viral CA may play a direct role in choosing integration sites or whether viral CA is acting behind the scenes to direct virus integration. Surely, new single-cell cutting-edge technologies are allowing and will continue to allow to build a new model of HIV-1 early steps (Figure 1).
References
- Barre-Sinoussi, F.; Chermann, J.C.; Rey, F.; Nugeyre, M.T.; Chamaret, S.; Gruest, J.; Dauguet, C.; Axler-Blin, C.; Vezinet-Brun, F.; Rouzioux, C.; et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 1983, 220, 868–871.
- Temin, H.M.; Mizutani, S. Viral RNA-dependent DNA Polymerase: RNA-dependent DNA Polymerase in Virions of Rous Sarcoma Virus. Nat. Cell Biol. 1970, 226, 1211–1213.
- Baltimore, D. Viral RNA-dependent DNA Polymerase: RNA-dependent DNA Polymerase in Virions of RNA Tumour Viruses. Nat. Cell Biol. 1970, 226, 1209–1211.
- Maldarelli, F.; Wu, X.; Su, L.; Simonetti, F.R.; Shao, W.; Hill, S.; Spindler, J.; Ferris, A.L.; Mellors, J.W.; Kearney, M.F.; et al. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 2014, 345, 179–183.
- Ezzell, C. AZT given the green light for clinical treatment of AIDS. Nat. Cell Biol. 1987, 326, 430.
- Hazuda, D.J.; Felock, P.; Witmer, M.; Wolfe, A.; Stillmock, K.; Grobler, J.A.; Espeseth, A.; Gabryelski, L.; Schleif, W.; Blau, C.; et al. Inhibitors of Strand Transfer That Prevent Integration and Inhibit HIV-1 Replication in Cells. Science 2000, 287, 646–650.
- Di Nunzio, F. New insights in the role of nucleoporins: A bridge leading to concerted steps from HIV-1 nuclear entry until integration. Virus Res. 2013, 178, 187–196.
- Berry, F.; Khalfi, P.; Maillot, F.; Seigneres, P.; Sid Ahmed, S.; Di Nunzio, F. Host nuclear pore factors: Team players of HIV-1 nuclear translocation and integration. Med. Sci. 2018, 34, 512–515.
- Fassati, A. Multiple roles of the capsid protein in the early steps of HIV-1 infection. Virus Res. 2012, 170, 15–24.
- Ambrose, Z.; Aiken, C. HIV-1 uncoating: Connection to nuclear entry and regulation by host proteins. Virology 2014, 455, 371–379.
- Campbell, E.M.; Hope, T.J. HIV-1 capsid: The multifaceted key player in HIV-1 infection. Nat. Rev. Genet. 2015, 13, 471–483.
- Novikova, M.; Zhang, Y.; Freed, E.O.; Peng, K. Multiple Roles of HIV-1 Capsid during the Virus Replication Cycle. Virol. Sin. 2019, 34, 119–134.
- Yant, S.R.; Mulato, A.; Hansen, D.; Tse, W.C.; Niedziela-Majka, A.; Zhang, J.R.; Stepan, G.J.; Jin, D.; Wong, M.H.; Perreira, J.M.; et al. A highly potent long-acting small-molecule HIV-1 capsid inhibitor with efficacy in a humanized mouse model. Nat. Med. 2019, 25, 1377–1384.
- Link, J.O.; Rhee, M.S.; Tse, W.C.; Zheng, J.; Somoza, J.R.; Rowe, W.; Begley, R.; Chiu, A.; Mulato, A.; Hansen, D.; et al. Clinical targeting of HIV capsid protein with a long-acting small molecule. Nat. Cell Biol. 2020, 584, 614–618.
- Bukrinsky, M.I.; Sharova, N.; McDonald, T.L.; Pushkarskaya, T.; Tarpley, W.G.; Stevenson, M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 1993, 90, 6125–6129.
- Farnet, C.M.; Haseltine, W.A. Determination of viral proteins present in the human immuno-deficiency virus type 1 preintegration complex. J. Virol. 1991, 65, 1910–1915.
- Blanco-Rodriguez, G.; Gazi, A.; Monel, B.; Frabetti, S.; Scoca, V.; Mueller, F.; Schwartz, O.; Krijnse-Locker, J.; Charneau, P.; Di Nunzio, F. Remodeling of the Core Leads HIV-1 Preintegration Complex into the Nucleus of Human Lymphocytes. J. Virol. 2020, 94.
- Fassati, A.; Goff, S.P. Characterization of Intracellular Reverse Transcription Complexes of Human Immunodeficiency Virus Type 1. J. Virol. 2001, 75, 3626–3635.
- Di Nunzio, F.; Fricke, T.; Miccio, A.; Valle-Casuso, J.C.; Perez, P.; Souque, P.; Rizzi, E.; Severgnini, M.; Mavilio, F.; Charneau, P.; et al. Nup153 and Nup98 bind the HIV-1 core and contribute to the early steps of HIV-1 replication. Virology 2013, 440, 8–18.
- Burdick, R.C.; Li, C.; Munshi, M.; Rawson, J.M.O.; Nagashima, K.; Hu, W.S.; Pathak, V.K. HIV-1 un-coats in the nucleus near sites of integration. Proc. Natl. Acad. Sci. USA 2020, 117, 5486–5493.
- Lee, K.; Ambrose, Z.; Martin, T.D.; Oztop, I.; Mulky, A.; Julias, J.G.; Vandegraaff, N.; Baumann, J.G.; Wang, R.; Yuen, W.; et al. Flexible Use of Nuclear Import Pathways by HIV-1. Cell Host Microbe 2010, 7, 221–233.
- Francis, A.C.; Marin, M.; Singh, P.K.; Achuthan, V.; Prellberg, M.J.; Palermino-Rowland, K.; Lan, S.; Tedbury, P.R.; Sarafianos, S.G.; Engelman, A.N.; et al. HIV-1 replication complexes accumulate in nuclear speckles and integrate into speckle-associated genomic domains. Nat. Commun. 2020, 11, 3505.
- Buffone, C.; Martinez-Lopez, A.; Fricke, T.; Opp, S.; Severgnini, M.; Cifola, I.; Petiti, L.; Frabetti, S.; Skorupka, K.; Zadrozny, K.K.; et al. Nup153 Unlocks the Nuclear Pore Complex for HIV-1 Nuclear Translocation in Nondividing Cells. J. Virol. 2018, 92.
- Achuthan, V.; Perreira, J.M.; Sowd, G.A.; Puray-Chavez, M.; McDougall, W.M.; Paulucci-Holthauzen, A.; Wu, X.; Fadel, H.J.; Poeschla, E.M.; Multani, A.S.; et al. Capsid-CPSF6 Interaction Licenses Nuclear HIV-1 Trafficking to Sites of Viral DNA Integration. Cell Host Microbe 2018, 24, 392–404.
- Sowd, G.A.; Serrao, E.; Wang, H.; Wang, W.; Fadel, H.J.; Poeschla, E.M.; Engelman, A.N. A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc. Natl. Acad. Sci. USA 2016, 113, E1054–E1063.
- Koh, Y.; Wu, X.; Ferris, A.L.; Matreyek, K.A.; Smith, S.J.; Lee, K.; KewalRamani, V.N.; Hughes, S.H.; Engelman, A.N. Differential Effects of Human Immunodeficiency Virus Type 1 Capsid and Cellular Factors Nucleoporin 153 and LEDGF/p75 on the Efficiency and Specificity of Viral DNA Integration. J. Virol. 2012, 87, 648–658.
- Rensen, E.; Mueller, F.; Scoca, V.; Parmar, J.J.; Souque, P.; Zimmer, C.; Di Nunzio, F. Clustering and reverse transcription of HIV-1 genomes in nuclear niches of macrophages. EMBO J. 2021, 40, e105247.
- Scoca, V.; Louveaux, M.; Morin, R.; Ershov, D.; Tinevez, J.-Y.; Di Nunzio, F. Direct tracking of single proviruses reveals HIV-1/LEDGF complexes excluded from virus-induced membraneless organelles. bioRxiv 2020.
- Lamond, A.I.; Spector, D.L. Nuclear speckles: A model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 2003, 4, 605–612.
- Peterlin, B.M.; Price, D.H. Controlling the Elongation Phase of Transcription with P-TEFb. Mol. Cell 2006, 23, 297–305.
- Huang, S.; Spector, D.L. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J. Cell. Biol. 1996, 133, 719–732.
- Kim, J.; Venkata, N.C.; Gonzalez, G.A.H.; Khanna, N.; Belmont, A.S. Gene expression amplification by nuclear speckle association. J. Cell Biol. 2019, 219.
- Müller, T.G.; Zila, V.; Peters, K.; Schifferdecker, S.; Stanic, M.; Lucic, B.; Laketa, V.; Lusic, M.; Müller, B.; Kräusslich, H.-G. HIV-1 uncoating by release of viral cDNA from capsid-like structures in the nucleus of infected cells. bioRxiv 2020.
- Lahaye, X.; Gentili, M.; Silvin, A.; Conrad, C.; Picard, L.; Jouve, M.; Zueva, E.; Maurin, M.; Nadalin, F.; Knott, G.J.; et al. NONO Detects the Nuclear HIV Capsid to Promote cGAS-Mediated Innate Immune Activation. Cell 2018, 175, 488–501.




