
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Katharina Jonas | + 2655 word(s) | 2655 | 2020-04-24 04:28:52 | | | |
| 2 | Catherine Yang | Meta information modification | 2655 | 2020-05-13 06:03:03 | | | | |
| 3 | Catherine Yang | -9 word(s) | 2646 | 2020-10-29 03:47:33 | | |
Video Upload Options
The majority of the genome is transcribed into pieces of non-(protein) coding RNA, among which long non-coding RNAs (lncRNAs) constitute a large group of particularly versatile molecules that govern basic cellular processes including transcription, splicing, RNA stability, and translation. The frequent deregulation of numerous lncRNAs in cancer is known to contribute to virtually all hallmarks of cancer. The post-transcriptional regulation of lncRNAs is mediated by RNA-binding proteins (RBPs). Interestingly, RBPs themselves are commonly deregulated in cancer and could thus constitute a major contribution to the deregulation of cancer-associated lncRNAs. Discussed here are four examples of well-known RBPs that regulate the transport or localization of cancer-associated lncRNAs and thereby impact the functionality of these lncRNAs. So far, out of the vast number of RBPs that exist, only a relatively small number has been found to specifically guide the transport or localization of cancer-related lncRNAs. In general, there is still a lack of knowledge about how lncRNAs are shuttled between or retained within different cellular compartments and future research will have to shed more light on these regulatory mechanisms.
1. Introduction
The concept that RNA merely serves as an intermediate, conveying the genetic information encoded in the form of DNA to be translated into proteins, has long been overthrown [1][2]. In fact, the vast majority of the genome does not code for proteins but is transcribed into various types of so-called non-coding RNAs (ncRNAs), which by themselves fulfill a multitude of pivotal regulatory functions [2][3][4][5]. Among these ncRNAs, one large group with particularly versatile functions are the long non-coding RNAs (lncRNAs), a class of ncRNAs that are defined as being longer than 200 nucleotides [6]. These lncRNAs can originate from different genomic locations. Most are interspersed between protein-coding genes (long intergenic non-coding RNAs, lincRNAs), while others are transcribed from the sense or antisense strands of introns and also exons of coding genes, and yet another type of lncRNAs originates from enhancer regions (eRNA) [7][8][9].
Compared to protein-coding genes, lncRNAs are poorly conserved between different species and their expression levels are rather low [7][8]. Initially, this led to the belief that they were nothing but transcriptional noise [6][7][8]. Soon, however, it was discovered that lncRNAs do exhibit considerable functionality, for example as regulators of transcription, but also on the post-transcriptional level by regulating mRNA splicing, stability and translation [6][7][8]. LncRNAs can also act as so-called competitive endogenous RNAs (ceRNAs), which sponge up microRNAs (miRNAs) and thus prevent them from binding to and inducing degradation or translational repression of their mRNA targets [10][11][12].
For a lncRNA to fulfill its function it is not only important how much of this lncRNA is present in a cell but equally, or potentially even more important, whether the lncRNA is properly transported to and located at its site of action. While lncRNAs are frequently enriched in the nucleus, they can also be located in the cytoplasm or in mitochondria and can be shuttled between these different compartments in response to different cellular conditions [8][13][14][15]. The transport of lncRNAs constitutes a form of post-transcriptional regulation that is mediated by RNA-binding proteins (RBPs) [16][17]. There is, however, not a lot known about how this transport of lncRNAs is facilitated [13][14]. Discussed below are four examples of well-known RBPs that have been found to regulate the transport or localization of cancer-associated lncRNAs. Importantly, these RBPs are themselves commonly deregulated in cancer and thus potentially contribute to the deregulation of lncRNAs in cancer.
2. Human Antigen R (HuR)
HuR, the protein product of the ELAV1 gene, is a ubiquitously expressed RBP that contains three RNA recognition motifs (RRMs) via which it preferentially binds to adenylate/uridylate-rich RNA elements (AREs) [18][19][20]. AREs are signals for rapid RNA degradation, and by blocking these recognition sites HuR can stabilize its RNA interaction partners [18][19][20]. HuR is frequently upregulated in cancer cells and is known to be involved in many hallmarks of cancer, such as invasion, angiogenesis, and inflammation, by post-transcriptionally regulating various cancer-related mRNAs [18][19][21][22][23]. In addition, HuR is known to not only regulate the level of lncRNAs by affecting their stability but has also been found to have an impact on lncRNA localization [24][25][26][27][28]. The different regulatory mechanisms that HuR exerts on its lncRNA targets are illustrated in Figure 1.
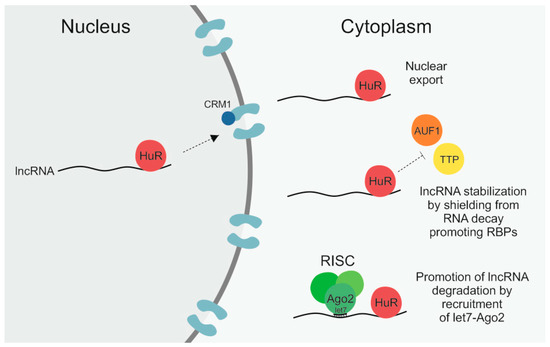
Figure 1. The RNA-binding protein (RBP) human antigen R (HuR) can regulate long non-coding RNAs (lncRNAs) by different means. First, by binding to its target lncRNA in the nucleus it can facilitate its subsequent nuclear export by interaction with the importin β superfamily member chromosomal maintenance 1 (CRM1). Secondly, by shielding lncRNAs from RNA decay promoting RBPs like ARE/poly(U)-binding/degradation factor 1 (AUF1) and tristetraprolin (TTP), HuR can enhance lncRNA stability. Thirdly, by recruitment of let7–Argonaute-2 (Ago2) it can promote lncRNA degradation via the RNA-induced silencing complex (RISC).
A study by Noh et al. found HuR to bind to both the 3′ and 5′ end of the RNA component of mitochondrial RNA processing endoribonuclease (RMRP), a 265-nt-long lncRNA that is best known as a component of the mitochondrial RNA-processing endoribonuclease (RNase MRP) complex and that is also involved in mitochondrial DNA replication [27][14]. Rather recently, an additional role of RMRP in different types of cancer has been reported by multiple publications that describe a contribution of RMRP to, for example, cancer cell proliferation, migration, and invasion, by sponging up different miRNAs [29][30][31][32]. According to Noh and colleagues, HuR binds to RMRP already in the nucleus and subsequently facilitates its nuclear export [27]. HuR itself can shuttle back and forth between the nucleus and cytoplasm in response to different stimuli [18][19]. The export of HuR from the nucleus occurs in a chromosomal maintenance 1 (CRM1)-dependent manner [27][33]. CRM1 is a member of the importin β superfamily that facilitates the nuclear export of proteins, to which it can bind either directly or via adaptors, as well as of RNA, in which case adaptor proteins are necessary [13][34]. Noh et al. showed that not only silencing of HuR itself in HEK293 cells but also of CRM1 led to significantly reduced cytoplasmic levels of nascent RMRP [27]. Importantly, there was no additive effect when both HuR and CRM1 were knocked down, highlighting that the nuclear export of RMRP was facilitated via the HuR–CRM1 axis [27]. While affecting the localization of RMRP, HuR was not observed to have an impact on the steady-state levels, meaning the stability, of the lncRNA [27]. There are a number of cancer-related lncRNAs whose stability is regulated by HuR, for example NEAT1 and lncRNA-p21 [24][25][26][28][35] and based on the high frequency of HuR binding sites, particularly in intronic regions [36][37], it can be anticipated that more will be identified in the future. However, so far, RMRP is the only lncRNA whose nuclear export has been reported to be mediated by HuR.
3. G-Rich RNA Sequence-Binding Factor 1 (GRSF1)
Even though the lncRNA RMRP is encoded in the nucleus, it is also present in mitochondria [27][14]. As detailed above, Noh et al. uncovered that the first step in the transport pathway of RMRP, the export from the nucleus into the cytoplasm, is facilitated by the RBP HuR [27]. In the same study the authors also reported that the subsequent import of RMRP into mitochondria seemed to be mediated via two import machineries, the TOM/TIM machinery and polynucleotide phosphorylase (PNPase) [27]. The TOM/TIM machinery consists of a translocase complex of the outer mitochondrial membrane (TOM) and two different inner membrane translocases (TIM) that together enable the transport of proteins across both mitochondrial membranes into the matrix [38]. The PNPase is located in the mitochondrial intermembrane space and promotes the import of RNA from the cytoplasm into the mitochondrial matrix [27][39]. How exactly RMRP is transported by these machineries and which RBPs it interacts with in order to do so has not been elucidated [27]. What Noh et al. did report was that the RBP GRSF1, while not directly contributing to its import, promoted the accumulation of RMRP in the mitochondrial matrix [27]. GRSF1 contains three RRMs via which it binds to a G-rich recognition motif (AGGGGD, with D = A/U), subsequently regulating splicing, polyadenylation, and export of its RNA targets [27][14][40]. One isoform of GRSF1 is located in mitochondria where it forms granules with newly synthesized mitochondrial RNAs, among them the two mitochondrial lncRNAs lncCyt b and lncND5, which carry 10 and 21 GRSF1 consensus-binding sites, respectively [14][40]. Knockdown of GRSF1 in immortalized primary fibroblasts decreased the overall level of these two mitochondrial lncRNAs by about half [40]. Noh et al., however, reported that the whole-cell level of RMRP was not affected by GRSF1 knockdown, but only its accumulation in the mitochondrial matrix [27]. Hence, GRSF1 seems to retain RMRP in the matrix once it has been imported, thereby facilitating its enrichment at its specific site of action [27].
RMRP is not the only example of a nuclear-encoded lncRNA that can be transported into mitochondria. The cancer-associated lncRNA MALAT1 (Metastasis-associated lung adenocarcinoma transcript 1) was also found to be present in mitochondria [15]. Under normal conditions, MALAT1 is mostly located in the nucleus where it regulates alternative splicing and gene expression [15][41][42]. In hepatocellular carcinoma (HCC), however, Zhao et al. observed an increased level of MALAT1 within mitochondria [15]. An RNA-fluorescent in situ hybridization (FISH) assay combined with mitochondrial staining showed that MALAT1 was highly enriched in HepG2 mitochondria but barely detectable in non-cancerous hepatic cells [15]. This mitochondrial accumulation of the lncRNA in HCC appears to enhance mitochondrial energy metabolism, thus potentially contributing to the oncogenic effects that MALAT1 exerts in HCC [15][43]. Interestingly, the study observed that for the mitochondrial lncRNA lncCyt b the situation is the other way around, namely that in non-cancerous cells it is located primarily in the mitochondrial matrix, where it is bound by GRSF1, but in HepG2 cells, it shows increased presence in the nucleus [15]. How the shuttling of these two lncRNAs between the nucleus and mitochondria is mediated is not known so far. It is clear, however, that different RBPs, like HuR and GRSF1, must play a central role in this process and that deregulations in this network of RBPs are responsible for the aberrant localization of lncRNAs in cancer, as observed for MALAT1 and lncCyt b in HCC. In case of lncCyt b, a disturbed interaction with GRSF1 could be one factor contributing to this phenomenon.
4. Insulin-Like Growth Factor 2 mRNA-Binding Protein 1 (IGF2BP1)
IGF2BP1, together with IGF2BP2 and IGF2BP3, belongs to a family of RBPs that is expressed in embryonic tissue and frequently reactivated in different types of cancer but rarely expressed in normal adult tissue [44][45]. Due to its regulation of stability, localization, and translation of numerous mRNAs like c-Myc [46][47], beta-catenin [48], and KRAS [49], it plays a role in embryogenesis and tumor development [50][51]. IGF2BP1 carries two RRMs in its N-terminal region and four hnRNP-K homology (KH) domains in the C-terminus, which are primarily responsible for its interaction with RNA targets in an N6-methyladenosine (m6A)-dependent manner [50][51]. The m6A modification is the most common type of RNA modification both in mRNAs and lncRNAs and stems from the addition of a methyl group to the N-6 position of adenosine by proteins like methyltransferase-like 3 (METTL3), METTL14, and Wilms’ tumor 1-associating protein (WTAP) [52][53].
IGF2BP1 is mostly cytoplasmic where it forms, together with its RNA targets and other RBPs, so-called messenger ribonucleoprotein (mRNP) granules [50][51][44][45][54]. When not associated with these mRNPs, IGF2BP1 can also translocate to the nucleus where it has been found to bind to mRNA already during transcription [50][51]. As IGF2BP1 contains two nuclear export signals (NES) within its second and fourth RNA-binding KH domain it is subsequently exported back into the cytoplasm together with its bound RNA target [53][54]. Hence, IGF2BP1 facilitates the nuclear export of its RNA target [53][54]. The prime example of this is given by beta-actin. IGF2BP1 binds to the beta-actin mRNA as soon as it is transcribed and subsequently enables its export into the cytoplasm, where IGF2BP2 either locates to perinuclear regions or interacts with and moves along the cytoskeleton towards the cell periphery, more precisely towards newly forming lamellipodia [52][54][55]. Here, IGF2BP2 is phosphorylated by the Src-kinase, which results in the disassociation of the beta-actin mRNA and its localized translation [55].
Following the same mechanism as observed for the beta-actin mRNA, IGF2BP1 also regulates the subcytoplasmic distribution of the lncRNA H19 [56]. H19 is a 2.3-kb-long, spliced and polyadenylated lncRNA that is, similarly to IGF2BP1, expressed during embryonic development and reactivated in several types of cancer [56][48][49][57]. H19 shows a diverse range of actions: it functions as a ceRNA, induces or represses the transcription of various genes, and interacts with and thus modulates the activity of different proteins like p53, thereby contributing to all hallmarks of cancer [57][58]. Runge et al. discovered that IGF2BP1 binds to the 3′ end of H19 with high affinity and thereby targets the lncRNA to lamellipodia and perinuclear regions of proliferating mouse embryonic fibroblasts [56]. In growth-arrested confluent cells, the IGF2BP1–H19 complex was dispersed more evenly in the cytoplasm [56]. It has been discovered that H19 seems to contribute to the migratory behavior and branching morphogenesis of epithelial cells, processes that are essential during embryogenic development but that also enable migration, invasion, and metastasis of cancerous cells [56][59]. This function of H19 coincides well with the targeting of IGF2BP1-H19 to the leading edge of proliferative cells.
5. Heterogeneous Nuclear Ribonucleoprotein K (hnRNPK)
The above discussed examples of RBPs are involved in the transport or localization of cancer-associated lncRNAs outside of the nucleus. In general, lncRNAs, however, tend to be enriched in nuclear fractions [8]. A well-studied RBP that plays a role in the nuclear accumulation of lncRNAs is the heterogeneous nuclear ribonucleoprotein K (hnRNPK) [60]. The exact mechanism of how hnRNPK can retain RNAs in the nucleus is however still unknown [60]. HnRNPK fulfills a wide variety of cellular functions, for example by acting as a transcription factor, regulating translation, and serving as a hub for various signaling pathways [61]. Numerous studies have observed oncogenic effects as well as a prognostic relevance of hnRNPK in different types of cancer, like breast, colorectal, and gastric cancer, where its overall levels are increased and it is aberrantly localized in the cytoplasm [62][63][64]. The binding of hnRNPK to RNA occurs via the interaction of its three KH domains, which are a type of RNA-binding motif first discovered in and therefore named after the RBP, with poly-C sequences in the RNA targets [65][66]. Lubelsky and Ulitsky, by screening a library of short fragments from nuclear mRNAs and lncRNAs, discovered a specific consensus sequence that is bound by hnRNPK and mediates nuclear accumulation [60]. This sequence consists of a 42-nt-long fragment that overlaps with Alu repeats, a very common type of short interspersed element (SINE), in antisense orientation and that contains three stretches of at least six pyrimidines (C/T), with two of these stretches matching the consensus sequence RCCTCCC (R = A/G) [60]. They termed the sequence SINE-derived nuclear RNA LOcalizatIoN (SIRLOIN) [60].
SIRLOINs are found in 13.1% of human lncRNAs and 7.5% of mRNAs and contribute substantially to the nuclear enrichment of RNA transcripts [60]. An example of a cancer-associated lncRNA that contains a SIRLOIN and is retained in the nucleus by hnRNPK is MALAT1 [60]. As stated before, the nuclear accumulation of MALAT1 has, however, been found to be disturbed in HCC, where undefined reasons lead to an increased mitochondrial presence of the lncRNA [15]. A recent study by Nguyen et al. showed that the deletion of a SINE in MALAT1 and the hence disrupted interaction with hnRNPK resulted in a more frequent translocation to the cytoplasm [67]. This was accompanied by increased DNA damage and apoptosis due to the redistribution of a protein called transactive response DNA binding protein 43 kDa (TDP-43) to the cytoplasm along with MALAT1 to which it is bound [67]. Aberrant expression and localization of hnRNPK, commonly observed in cancer, could thus also play a role in the change of MALAT1 localization in HCC. As reviewed elsewhere, there are numerous examples for interactions between lncRNAs and hnRNPK where they jointly regulate gene expression by various means [65]. Thus, the deregulation of hnRNPK in cancer has far-reaching ramifications as it affects the function and localization of lncRNAs [65].
References
- Hui Ling; Kimberly Vincent; Martin Pichler; Riccardo Fodde; Ioana Berindan-Neagoe; Frank J. Slack; George A. Calin; Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene 2015, 34, 5003-5011, 10.1038/onc.2014.456.
- Eleni Anastasiadou; Alberto Faggioni; Pankaj Trivedi; Frank J. Slack; The Nefarious Nexus of Noncoding RNAs in Cancer. International Journal of Molecular Sciences 2018, 19, 2072, 10.3390/ijms19072072.
- Jason M. Johnson; Stephen Edwards; Daniel Shoemaker; Eric E. Schadt; Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends in Genetics 2005, 21, 93-102, 10.1016/j.tig.2004.12.009.
- Barbara L. Kee; A comprehensive transcriptional landscape of human hematopoiesis.. Cell Stem Cell 2011, 8, 122-4, 10.1016/j.stem.2011.01.006.
- Maria A. Smolle; Felix Prinz; George A. Calin; Martin Pichler; Current concepts of non-coding RNA regulation of immune checkpoints in cancer.. Molecular Aspects of Medicine 2019, 70, 117-126, 10.1016/j.mam.2019.09.007.
- Runwen Yao; Yang Wang; Ling-Ling Chen; Cellular functions of long noncoding RNAs.. Nature 2019, 21, 542-551, 10.1038/s41556-019-0311-8.
- Lina Ma; Vladimir B. Bajic; Zhang Zhang; On the classification of long non-coding RNAs.. RNA Biology 2013, 10, 925-33, 10.4161/rna.24604.
- Thomas Derrien; Rory Johnson; Giovanni Bussotti; Andrea Tanzer; Sarah Djebali; Hagen Tilgner; Gregory Guernec; David Martín; Angelika Merkel; David G. Knowles; Julien Lagarde; Lavanya Veeravalli; Xiaoan Ruan; Yijun Ruan; Timo Lassmann; Piero Carninci; James B. Brown; Leonard Lipovich; Jose M Gonzalez; Mark Thomas; Carrie A. Davis; Ramin Shiekhattar; Thomas R. Gingeras; Tim @timjph Hubbard; Cedric Notredame; Jennifer Harrow; Roderic Guigó; The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Research 2012, 22, 1775-1789, 10.1101/gr.132159.111.
- Preston R. Arnold; Andrew D. Wells; Xian C. Li; Diversity and Emerging Roles of Enhancer RNA in Regulation of Gene Expression and Cell Fate.. Frontiers in Cell and Developmental Biology 2020, 7, 377, 10.3389/fcell.2019.00377.
- Christiane Klec; Felix Prinz; Martin Pichler; Involvement of the long noncoding RNA NEAT1 in carcinogenesis.. Molecular Oncology 2018, 13, 46-60, 10.1002/1878-0261.12404.
- Chen Wang; Xiaoxue Jiang; Xiaonan Li; Shuting Song; Qiuyu Meng; Liyan Wang; Yanan Lu; Xiaoru Xin; Hu Pu; Xin Gui; Tianming Li; Dongdong Lu; Long noncoding RNA HULC accelerates the growth of human liver cancer stem cells by upregulating CyclinD1 through miR675-PKM2 pathway via autophagy.. Stem Cell Research & Therapy 2020, 11, 8-14, 10.1186/s13287-019-1528-y.
- Hui Zhou; Zixu Gao; Fusheng Wan; Taurine-upregulated gene 1 contributes to cancers through sponging microRNA.. Acta Biochimica et Biophysica Sinica 2019, 51, 123-130, 10.1093/abbs/gmy156.
- Tobias Williams; Linh H. Ngo; Vihandha O. Wickramasinghe; Nuclear export of RNA: Different sizes, shapes and functions. Seminars in Cell & Developmental Biology 2018, 75, 70-77, 10.1016/j.semcdb.2017.08.054.
- Yaru Dong; Takeshi Yoshitomi; Ji-Fan Hu; Jizhe Cui; Long noncoding RNAs coordinate functions between mitochondria and the nucleus. Epigenetics & Chromatin 2017, 10, 41, 10.1186/s13072-017-0149-x.
- Yijing Zhao; Shanshan Liu; Lei Zhou; Xueli Li; Ying Meng; Yan Li; Lingyu Li; Benzheng Jiao; Ling Bai; Yu Yu; Songling Zhang; Wei Li; Andrew R. Hoffman; Ji-Fan Hu; Jiuwei Cui; Aberrant shuttling of long noncoding RNAs during the mitochondria-nuclear crosstalk in hepatocellular carcinoma cells.. American journal of cancer research 2019, 9, 999-1008, null.
- Shakur Mohibi; Xinbin Chen; Jin Zhang; Cancer the‘RBP’eutics–RNA-binding proteins as therapeutic targets for cancer. Pharmacology & Therapeutics 2019, 203, 107390, 10.1016/j.pharmthera.2019.07.001.
- Stefanie Gerstberger; Markus Hafner; Thomas Tuschl; A census of human RNA-binding proteins. Nature Reviews Genetics 2014, 15, 829-845, 10.1038/nrg3813.
- Schultz, C.W.; Preet, R.; Dhir, T.; Dixon, D.A.; Brody, J.R. Understanding and targeting the disease-related RNA binding protein human antigen R (HuR). WIREs RNA 2020, 11, e1581, doi:10.1002/wrna.1581.
- Jun Wang; Yan Guo; Huili Chu; Yaping Guan; Jingwang Bi; Baocheng Wang; Multiple Functions of the RNA-Binding Protein HuR in Cancer Progression, Treatment Responses and Prognosis. International Journal of Molecular Sciences 2013, 14, 10015-10041, 10.3390/ijms140510015.
- Nina Ripin; Julien Boudet; Malgorzata M Duszczyk; Alexandra Hinniger; Michael Faller; Miroslav Krepl; Abhilash Gadi; Robert Schneider; Judit Sponer; Nicole C. Meisner-Kober; Frédéric H.-T. Allain; Molecular basis for AU-rich element recognition and dimerization by the HuR C-terminal RRM. Proceedings of the National Academy of Sciences 2019, 116, 2935-2944, 10.1073/pnas.1808696116.
- Cecilia Osera; Jennifer L. Martindale; Marialaura Amadio; Jiyoung Kim; Xiaoling Yang; Christopher A. Moad; Fred E. Indig; Stefano Govoni; Kotb Abdelmohsen; Myriam Gorospe; Alessia Pascale; Induction ofVEGFAmRNA translation by CoCl2mediated by HuR. RNA Biology 2015, 12, 1121-1130, 10.1080/15476286.2015.1085276.
- Wei Xu; Chao Chen; Ruijun Xu; Yifan Li; Ruixi Hu; Zhikun Li; Xiaodong Zhu; Knockdown of HuR represses osteosarcoma cells migration, invasion and stemness through inhibition of YAP activation and increases susceptibility to chemotherapeutic agents. Biomedicine & Pharmacotherapy 2018, 102, 587-593, 10.1016/j.biopha.2018.03.098.
- Weidan Peng; Narumi Furuuchi; Ludmila Aslanukova; Yu-Hung Huang; Samantha Z. Brown; Wei Jiang; Sankar Addya; Vikalp Vishwakarma; Erika Peters; Jonathan R. Brody; Dan A. Dixon; Janet A. Sawicki; Elevated HuR in Pancreas Promotes a Pancreatitis-Like Inflammatory Microenvironment That Facilitates Tumor Development. Molecular and Cellular Biology 2018, 38, e00427-17, 10.1128/MCB.00427-17.
- Yun-Ping Hu; Yun-Peng Jin; Xiang-Song Wu; Yang Yang; Yong-Sheng Li; Huai-Feng Li; Shan-Shan Xiang; Xiao-Ling Song; Lin Jiang; Yi-Jian Zhang; Wen Huang; Shi-Li Chen; Fa-Tao Liu; Chen Chen; Qin Zhu; Hongzhuan Chen; Rong Shao; Yingbin Liu; LncRNA-HGBC stabilized by HuR promotes gallbladder cancer progression by regulating miR-502-3p/SET/AKT axis.. Molecular Cancer 2019, 18, 167-18, 10.1186/s12943-019-1097-9.
- Je-Hyun Yoon; Kotb Abdelmohsen; Subramanya Srikantan; Xiaoling Yang; Jennifer L. Martindale; Supriyo De; Maite Huarte; Ming Zhan; Kevin G. Becker; Myriam Gorospe; LincRNA-p21 Suppresses Target mRNA Translation. Molecular Cell 2013, 50, 303, 10.1016/j.molcel.2013.04.008.
- Je-Hyun Yoon; Kotb Abdelmohsen; Jiyoung Kim; Xiaoling Yang; Jennifer L. Martindale; Kumiko Tominaga-Yamanaka; Elizabeth J. White; Arturo V. Orjalo; John L. Rinn; Stefan G. Kreft; Gerald Wilson; Myriam Gorospe; Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination.. Nature Communications 2013, 4, 2939-2939, 10.1038/ncomms3939.
- Ji Heon Noh; Kyoung Mi Kim; Kotb Abdelmohsen; Je-Hyun Yoon; Amaresh C. Panda; Rachel Munk; Jiyoung Kim; Jessica Curtis; Christopher A. Moad; Christina M. Wohler; Fred E. Indig; Wilson De Paula; Dawood B. Dudekula; Supriyo De; Yulan Piao; Xiaoling Yang; Jennifer L. Martindale; R. De Cabo; Myriam Gorospe; HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes & Development 2016, 30, 1224-1239, 10.1101/gad.276022.115.
- Wei Lei; Zhi-Long Wang; He-Jun Feng; Xiang-Dan Lin; Chuang-Zhong Li; Di Fan; Long non-coding RNA SNHG12promotes the proliferation and migration of glioma cells by binding to HuR. International Journal of Oncology 2018, null, , 10.3892/ijo.2018.4478.
- Ning Zhou; Zili He; Hongying Tang; Bo Jiang; Wei Cheng; LncRNA RMRP/miR-613 axis is associated with poor prognosis and enhances the tumorigenesis of hepatocellular carcinoma by impacting oncogenic phenotypes.. American journal of translational research 2019, 11, 2801-2815, null.
- Lingyu Tang; Yuting Wang; Huishan Wang; Boming Xu; Hao Ji; Guolong Xu; Xianxiu Ge; Quanpeng Li; Lin Miao; Long noncoding-RNA component of mitochondrial RNA processing endoribonuclease is involved in the progression of cholangiocarcinoma by regulating microRNA-217.. Cancer Science 2019, 110, 2166-2179, 10.1111/cas.14074.
- Xichun Xiao; Yueli Gu; Genjie Wang; Shuxia Chen; c-Myc, RMRP, and miR-34a-5p form a positive-feedback loop to regulate cell proliferation and apoptosis in multiple myeloma. International Journal of Biological Macromolecules 2019, 122, 526-537, 10.1016/j.ijbiomac.2018.10.207.
- H-L Cao; Z-J Liu; P-L Huang; Y-L Yue; J-N Xi; lncRNA-RMRP promotes proliferation, migration and invasion of bladder cancer via miR-206.. European review for medical and pharmacological sciences 2019, 23, 1012-1021, null.
- Christopher M. Brennan; Imed-Eddine Gallouzi; Joan A. Steitz; Protein Ligands to Hur Modulate Its Interaction with Target Mrnas in Vivo. Journal of Cell Biology 2000, 151, 1-14, 10.1083/jcb.151.1.1.
- Kevin T Nguyen; Michael P Holloway; Rachel A. Altura; The CRM1 nuclear export protein in normal development and disease. International journal of biochemistry and molecular biology 2012, 3, 137-151, null.
- Yiqing Chai; Jie Liu; Zhikun Zhang; Liwei Liu; HuR-regulated lncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer. Cancer Medicine 2016, 5, 1588-1598, 10.1002/cam4.710.
- Tala Bakheet; Edward Hitti; Maher Al-Saif; Walid N. Moghrabi; Khalid S. A. Khabar; The AU-rich element landscape across human transcriptome reveals a large proportion in introns and regulation by ELAVL1/HuR.. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer 2018, 1861, 167-177, 10.1016/j.bbagrm.2017.12.006.
- Marvin Jens; Transcriptome-Wide Analysis of Regulatory Interactions of the RNA-Binding Protein HuR. Springer Theses 2014, null, 35-54, 10.1007/978-3-319-07082-7_3.
- Carla M. Koehler; Protein translocation pathways of the mitochondrion.. FEBS Letters 2000, 476, 27-31, 10.1016/s0014-5793(00)01664-1.
- Geng Wang; Hsiao-Wen Chen; Yavuz Oktay; Jin Zhang; Eric L. Allen; Geoffrey M. Smith; Kelly C. Fan; Jason S. Hong; Samuel W. French; J. Michael McCaffery; Robert N. Lightowlers; Herbert C. Morse; Carla M. Koehler; Michael A. Teitell; PNPASE Regulates RNA Import into Mitochondria. Cell 2010, 142, 456-67, 10.1016/j.cell.2010.06.035.
- Hana Antonicka; Florin Sasarman; Tamiko Nishimura; Vincent Paupe; Eric A Shoubridge; The Mitochondrial RNA-Binding Protein GRSF1 Localizes to RNA Granules and Is Required for Posttranscriptional Mitochondrial Gene Expression. Cell Metabolism 2013, 17, 386-398, 10.1016/j.cmet.2013.02.006.
- Tony Gutschner; Monika Hämmerle; Sven Diederichs; Monika Haemmerle; MALAT1 — a paradigm for long noncoding RNA function in cancer. Journal of Molecular Medicine 2013, 91, 791-801, 10.1007/s00109-013-1028-y.
- Vidisha Tripathi; Jonathan D. Ellis; Zhen Shen; David Y. Song; Qun Pan; Andrew T. Watt; Susan M. Freier; C. Frank Bennett; Alok Sharma; Paula A. Bubulya; Benjamin J. Blencowe; Supriya G. Prasanth; Kannanganattu V. Prasanth; The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Molecular Cell 2010, 39, 925-38, 10.1016/j.molcel.2010.08.011.
- Maryam Abbastabar; Mohammad Sarfi; Abolfazl Golestani; Ehsan Khalili; lncRNA involvement in hepatocellular carcinoma metastasis and prognosis. EXCLI journal 2018, 17, 900-913, 10.17179/excli2018-1541.
- Gu, W.; Wells, A.L.; Pan, F.; Singer, R.H. Feedback regulation between zipcode binding protein 1 and beta-catenin mRNAs in breast cancer cells. Mol. Cell. Biol. 2008, 28, 4963–4974, doi:10.1128/MCB.00266-08.
- Perry S. Mongroo; Felicite K. Noubissi; Miriam Cuatrecasas; Jiri Kalabis; Catrina E. King; Cameron N. Johnstone; Mark J. Bowser; Antoni Castells; Vladimir S. Spiegelman; Anil K. Rustgi; IMP-1 displays cross-talk with K-Ras and modulates colon cancer cell survival through the novel proapoptotic protein CYFIP2.. Cancer Research 2011, 71, 2172-82, 10.1158/0008-5472.CAN-10-3295.
- Lina Liu; Yuwei Wang; Jie Wu; Jingwen Liu; Zongchang Qin; Hong Fan; N6-Methyladenosine: A Potential Breakthrough for Human Cancer. Molecular Therapy - Nucleic Acids 2020, 19, 804-813, 10.1016/j.omtn.2019.12.013.
- Sihui Yu; Xi Li; Shiyun Liu; Rui Yang; Xiangnan Liu; Sufang Wu; N6-Methyladenosine: A Novel RNA Imprint in Human Cancer.. Frontiers in Oncology 2019, 9, 1407, 10.3389/fonc.2019.01407.
- Xinwei Huang; Hong Zhang; Xiaoran Guo; Zongxin Zhu; Haibo Cai; Xiangyang Kong; Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in cancer.. Journal of Hematology & Oncology 2018, 11, 88, 10.1186/s13045-018-0628-y.
- Bell, J.L.; Wächter, K.; Mühleck, B.; Pazaitis, N.; Köhn, M.; Lederer, M.; Hüttelmaier, S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): Post-transcriptional drivers of cancer progression? Cell. Mol. Life Sci. CMLS 2013, 70, 2657–2675, doi:10.1007/s00018-012-1186-z.
- Doreen Weidensdorfer; Nadine Stöhr; Anne Baude; Marcell Lederer; Marcel Köhn; Angelika Schierhorn; Sabine Buchmeier; Elmar Wahle; Stefan Hüttelmaier; Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA 2008, 15, 104-115, 10.1261/rna.1175909.
- Dan Sparanese; Chow H. Lee; CRD-BP shields c-myc and MDR-1 RNA from endonucleolytic attack by a mammalian endoribonuclease.. Nucleic Acids Research 2007, 35, 1209-21, 10.1093/nar/gkl1148.
- Finn C Nielsen; Jacob Nielsen; Mette A Kristensen; Grete Koch; Jan Christiansen; Cytoplasmic trafficking of IGF-II mRNA-binding protein by conserved KH domains.. Journal of Cell Science 2002, 115, , null.
- Jacob Nielsen; Sidsel K. Adolph; Ewa Rajpert-De Meyts; Jens Lykke-Andersen; Grete Koch; Jan Christiansen; Finn C. Nielsen; Nuclear transit of human zipcode-binding protein IMP1. Biochemical Journal 2003, 376, 383-391, 10.1042/bj20030943.
- Yuri Oleynikov; Robert H. Singer; Real-Time Visualization of ZBP1 Association with β-Actin mRNA during Transcription and Localization. Current Biology 2003, 13, 199-207, 10.1016/s0960-9822(03)00044-7.
- Biswas, J.; Nunez, L.; Das, S.; Yoon, Y.J.; Eliscovich, C.; Singer, R.H. Zipcode Binding Protein 1 (ZBP1; IGF2BP1): A Model for Sequence-Specific RNA Regulation. Cold Spring Harb. Symp. Quant. Biol. 2020, doi:10.1101/sqb.2019.84.039396.
- Steffen Runge; Finn Cilius Nielsen; Jacob Nielsen; Jens Lykke-Andersen; Ulla M. Wewer; Jan Christiansen; H19 RNA Binds Four Molecules of Insulin-like Growth Factor II mRNA-binding Protein. Journal of Biological Chemistry 2000, 275, 29562-29569, 10.1074/jbc.m001156200.
- Clément Lecerf; Xuefen Le Bourhis; Eric Adriaenssens; The long non-coding RNA H19: an active player with multiple facets to sustain the hallmarks of cancer. Cellular and Molecular Life Sciences 2019, 76, 4673-4687, 10.1007/s00018-019-03240-z.
- M.F. Roizen; Hallmarks of Cancer: The Next Generation. Yearbook of Anesthesiology and Pain Management 2012, 2012, 13, 10.1016/j.yane.2012.02.046.
- Eric Adriaenssens; Séverine Lottin; Nathalie Berteaux; Louis Hornez; William Fauquette; Véronique Fafeur; Jean-Philippe Peyrat; Xuefen Le Bourhis; Hubert Hondermarck; Jean Coll; Thierry Dugimont; Jean-Jacques Curgy; Cross-Talk between Mesenchyme and Epithelium Increases H19 Gene Expression during Scattering and Morphogenesis of Epithelial Cells. Experimental Cell Research 2002, 275, 215-229, 10.1006/excr.2002.5500.
- Yoav Lubelsky; Igor Ulitsky14; Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature 2018, 555, 107-111, 10.1038/nature25757.
- Ziyi Wang; Heng Qiu; Jianbo He; Langxia Liu; Wei Xue; Archa Fox; Jennifer Tickner; Jiake Xu; The emerging roles of hnRNPK. Journal of Cellular Physiology 2019, 235, 1995-2008, 10.1002/jcp.29186.
- Mahitosh Mandal; Ratna Vadlamudi; Diep Nguyen; Rui-An Wang; Luis Costa; Rozita Bagheri-Yarmand; John Mendelsohn; Rakesh Kumar; Growth Factors Regulate Heterogeneous Nuclear Ribonucleoprotein K Expression and Function. Journal of Biological Chemistry 2000, 276, 9699-9704, 10.1074/jbc.m008514200.
- B Carpenter; M McKay; S R Dundas; L C Lawrie; C Telfer; G I Murray; Heterogeneous nuclear ribonucleoprotein K is over expressed, aberrantly localised and is associated with poor prognosis in colorectal cancer. British Journal of Cancer 2006, 95, 921-927, 10.1038/sj.bjc.6603349.
- Ruirui Yang; Ying Zeng; Haifan Xu; Zhuo Chen; Mengqin Xiang; Yun Fu; Yufang Yin; Jing Zhong; Min Zeng; Peihua Wang; Qin You; Xi Zeng; Heterogeneous nuclear ribonucleoprotein K is overexpressed and associated with poor prognosis in gastric cancer. Oncology Reports 2016, 36, 929-935, 10.3892/or.2016.4845.
- Yongjie Xu; Wei Wu; Qiu Han; Yaling Wang; Cencen Li; Pengpeng Zhang; Haixia Xu; New Insights into the Interplay between Non-Coding RNAs and RNA-Binding Protein HnRNPK in Regulating Cellular Functions. Cells 2019, 8, 62, 10.3390/cells8010062.
- H Siomi; M J Matunis; W M Michael; G Dreyfuss; The pre-mRNA binding K protein contains a novel evolutionarily conserved motif.. Nucleic Acids Research 1993, 21, 1193-1198, null.
- Tuan M. Nguyen; Elena B. Kabotyanski; Lucas Reineke; Jiaofang Shao; Feng Xiong; Joo-Hyung Lee; Julien Dubrulle; Hannah Johnson; Fabio Stossi; Phoebe Tsoi; Kyoung-Jae Choi; Alexander G. Ellis; Na Zhao; Jin Cao; Oluwatoyosi Adewunmi; Josephine Ferreon; Allan Chris M. Ferreon; Joel R. Neilson; Michael A. Mancini; Xi Chen; Wenbo Li; Jeffrey M. Rosen; The SINEB1 Element in the Long Non-Coding RNA Malat1 is Necessary for TDP-43 Proteostasis. SSRN Electronic Journal 2018, null, , 10.2139/ssrn.3299453.




