
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Beow Keat Yap | + 4033 word(s) | 4033 | 2020-12-08 07:12:43 | | | |
| 2 | Dean Liu | -2492 word(s) | 1541 | 2021-01-25 07:34:03 | | |
Video Upload Options
14-3-3σ is an acidic homodimer protein with more than one hundred different protein partners associated with oncogenic signaling and cell cycle regulation.
1. Introduction
The 14-3-3 proteins are a group of acidic polypeptides that are highly conserved in all eukaryotic cells[1][2][3]. The 14-3-3 family was initially described by Moore & Perez in 1967 as an abundant mammalian brain protein family which took its name based on its elution profile, specifically the fraction number of bovine brain homogenate from diethylaminoethyl (DEAE) cellulose column (14th fraction) and subsequent purified fraction 3.3 from gel electrophoresis[4][5][6][7][8]. The 14-3-3 family comprises seven human isoforms which are named after their respective elution positions on high performance liquid chromatography (HPLC) (β-beta, ε-epsilon, γ-gamma, η-eta, σ-sigma, τ-tau, and ζ-zeta) with at least 500 partners forming protein–protein interaction (PPI) in mammalian cells[9][10][11][12]. Moreover, 14-3-3 proteins have also been detected in non-vertebrate species such as plants and yeasts [13][14][15][16][17]. The overall structure of 14-3-3 proteins is highly conserved among the family members with a molecular mass of approximately 28–30 kDa and isoelectric point of 4–5[9][18]. Crystal structures of 14-3-3 proteins revealed that they are highly helical with a clamp-like shape dimer. All human 14-3-3 isoforms are expressed as both homo- and heterodimers. The dimer form of 14-3-3 proteins is capable of binding two ligand motifs at the same time, either from the same target or from two different partners[19].
The 14-3-3 proteins are also classified as phosphoserine/phosphothreonine (pSer/pThr)-recognition proteins, as they generally exert their activity through binding to the phosphoserine/phosphothreonine-containing motifs of a multitude of molecules with various functions such as kinases, phosphatases, transmembrane receptors, and transcription factors[2][20][21][22]. In general, there are two high-affinity phosphorylation-dependent binding motifs that are recognized by the amphipathic binding grooves of all 14-3-3 isoforms, i.e., Arg-Ser-Xaa-pSer-Xaa-Pro (R-S-X-pS-X-P, mode I, Figure 1a) and Arg-Xaa-Xaa-Xaa-pSer/Thr-Xaa-Pro (R-X-X-X-pS/T-X-P, mode II, Figure 1b), where X is any amino acid and pS/T represents phosphorylated serine or threonine[23][24][25][26][27]. A third binding motif recognized by the C-terminus of 14-3-3 proteins, i.e., pS/pT-X1–2-COOH (mode III, Figure 1c) has also been reported [28][29]. Nevertheless, not all 14-3-3 interactions require a phosphorylated residue as 14-3-3 has also been reported to bind to several non-phosphorylated proteins and peptides, such as exoenzyme S, Cdc25B, and p190RhoGEF [30][31][32][33][34][35].
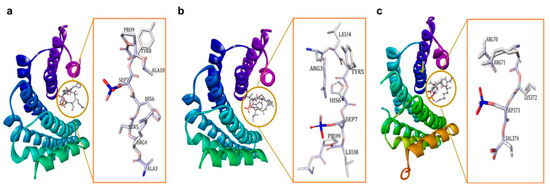
Figure 1. (a) 14-3-3ζ/phosphopeptide complex (mode 1, PDB: 1QJB), (b) 14-3-3ζ/phosphopeptide complex (mode II, PDB: 1QJA), (c) 14-3-3σ/TASK3 peptide (mode III, PDB: 6GHP).
Consistent with the ability of 14-3-3 proteins to bind to various binding motifs, 14-3-3 proteins are found to be involved in a wide range of physiological processes which include cell proliferation[36][37][38], cell cycle control [39][40][41][42][43], and cell apoptosis[44][45][46][47].
2. Role of 14-3-3σ in Cancer
The 14-3-3σ protein has attracted the attention of researchers as a vital target to fight against cancer growth and metastasis. Previous studies have demonstrated the role of 14-3-3σ in suppressing tumor metabolic reprogramming [48]. In addition, few reports have also highlighted the crucial role of 14-3-3σ against the cancer cell invasion and metastasis. For instance, a low level of 14-3-3σ has been shown to promote production of lactate which stimulates the migration of epithelial cancer cells to a distant organ through breaking down of extracellular matrix[48][49]. Studies have also showed that, among all seven well-known human 14-3-3 isoforms, 14-3-3σ is the only isoform that possesses tumor-suppressing activity[9][19][[50][51][52]. It has been shown that 14-3-3σ protein directly controls the G2-M checkpoint of the cell cycle by protecting the tumor suppressor factor P53 against the MDM2-mediated ubiquitination and degradation[53][54][55]. In addition, 14-3-3σ was also reported to play a crucial role in the cell cycle arrest regulation by acting as a cyclin-dependent kinase (Cdk) inhibitor, i.e., through sequestering the cyclin-dependent kinase 1-cyclin B1 complex from entering nucleus and initiate mitosis, as well as binding to the cyclin-dependent kinases 2 and 4[56][57]. Moreover, 14-3-3σ was also found to negatively regulates the oncogenic activity of the Protein kinase B (also known as Akt) and thus protecting against Akt-mediated tumorigenesis[53]. Further, 14-3-3σ has also been reported as a target gene in mammary epithelial cells which regulates the antiproliferative activity of the transforming growth factor-beta 1 (TGF-b1) through the Smad3-dependent mechanism[58][59]. Furthermore, reports have demonstrated 14-3-3σ involvement in controlling cell proliferation and cancer metastasis via the termination of NF-ĸB signal in mammary cells by regulating the nuclear export of the p65 subunit of NF-ĸB transcription factor and subsequently inhibits its transcriptional activity[60][61]. Moreover, 14-3-3σ has also been reported to regulate the expression of human TASK-3 channel (which is believed to facilitate cancer cell’s proliferation and survival), by blocking the endoplasmic reticulum retention sequences, and thereby promoting the surface expression of this channel[62][63][64]. 14-3-3σ also regulates the oncogenic activity of transcriptional coactivator TAZ which is an oncogenic protein that promotes cell proliferation and migration. The binding of TAZ to 14-3-3σ leads to cytoplasmic retention of TAZ which subsequently disabling its function[65][66].
Unlike other isoforms which show elevated expression in many types of cancer, 14-3-3σ protein level is downregulated in chronic myeloid leukaemia, nasopharyngeal carcinoma, as well as lung, breast, oesophageal, uterine, ovarian, and skin cancers[2][67][68][69][70][71]. The low expression level of 14-3-3σ protein in many cancer types has been linked to either promoter hypermethylation of Sfn gene (which encodes the 14-3-3σ protein) or direct 14-3-3σ degradation through ubiquitination which eventually aborts the normal physiological role of 14-3-3σ against tumor growth and metastasis[51][72][73][74][75]. Consistent with these observations, introduction of a DNA demethylating agent, 5-aza-20-deoxycytidine significantly upregulated the expression level of 14-3-3σ in salivary gland adenoid cystic carcinoma and nasopharyngeal carcinoma[76]. In addition, a separate study demonstrated that an upregulation of 14-3-3σ expression by Marsdenia tenacissima extract was able to mediate G2/M cell cycle arrest in breast cancer [78].
Although numerous studies have showed the vital role of 14-3-3σ in controlling the tumor formations and metastasis, some studies have also indicated that the 14-3-3σ could be a double-edged sword [68] as its upregulation has also been linked with resistance to chemotherapeutic agents[79][80][81]. In addition, studies have shown that 14-3-3σ also induces overexpression of matrix metalloproteinase 1 (MMP-1), a proteolytic enzyme that degrades native fibrillar collagens, and is often associated with poor prognosis in malignant tumor[62][82][83]. Furthermore, 14-3-3σ has also been reported to bind to the c-Abl protein, preventing its nuclear translocation and subsequently interfering with its pro-apoptotic effect[84][85].
3. Conclusions
In conclusion, the aberrant expression of 14-3-3σ has been observed in many cancers. Various protein partners and mechanisms involving 14-3-3σ in cancer growth and metastasis have been reported. This suggests that 14-3-3σ is an important target for anticancer drug discovery and development. Consistent with this observation, different chemical classes of 14-3-3σ PPI modulators have been developed as potential therapeutics against cancer. This includes 14-3-3σ PPI stabilizers such as fusicoccanes analogues and fragment-derived small molecule stabilizers, as well as phosphonate and non-phosphonate type 14-3-3σ PPI inhibitors. These modulators were successfully identified using a combination of techniques including in silico tools (ligand-based screening, docking, molecular dynamics simulations), biophysical techniques (NMR, X-ray crystallography, isothermal titration calorimetry), fluorescence polarization, as well as cell-based assays.
However, it is worth noting that both inhibitors and stabilizers of 14-3-3σ PPI available to date mainly target the amphipathic binding pocket. While inhibitors bind directly to the three key amino acids in the amphipathic binding pocket (Arg56, Arg129, and Tyr130), the stabilizers generally bind to the site adjacent to the amphipathic binding pocket, as the amphipathic binding pocket is often occupied by the protein partner of 14-3-3σ. Having said that, a direct interaction with Lys122 at the amphipathic binding pocket of 14-3-3σ was observed in both inhibitors and stabilizers. This suggests that a 14-3-3σ PPI inhibitor is also likely to interfere with the binding of other 14-3-3σ partners which are involved in suppressing cancer cell growth, metabolism, and metastasis, such as the tumor suppressor gene P53, TASK-3, p65, and TAZ. Intriguingly, these amino acid residues are also conserved among all 14-3-3 isoforms. This suggests that modulators that target the amphipathic binding groove of 14-3-3σ may also bind to other isoforms, and may produce other undesirable effects since only 14-3-3σ is frequently downregulated in cancer while other isoforms are usually upregulated.
Although the molecular tweezer seems promising as a potentially selective 14-3-3σ inhibitor as it has been reported to bind to the C-terminal domain of 14-3-3σ, rather than the amphipathic binding pocket, and yet is effective in displacing the binding of the protein partner from 14-3-3σ, it is still unclear if this inhibitor is indeed selective to 14-3-3σ since recent finding seems to suggest that molecular tweezer may binds to any solvently exposed Lys residues. Moreover, the interacting amino acid residue Lys214 is also conserved across all isoforms. Nevertheless, it is clearly demonstrated that it is possible to target other sites on 14-3-3σ in modulating its PPI interaction and is potentially the way forward for the design of new highly selective modulators of 14-3-3σ in the future.
References
- Ballone, A.; Centorrino, F.; Ottmann, C. 14-3-3: A case study in PPI modulation. Molecules 2018, 23, 1386.
- Fan, X.; Cui, L.; Zeng, Y.; Song, W.; Gaur, U.; Yang, M. 14-3-3 proteins are on the crossroads of cancer, aging, and age-related neurodegenerative disease. Int. J. Mol. Sci. 2019, 20, 3518.
- Tugaeva, K.V.; Titterington, J.; Sotnikov, D.V.; Maksimov, E.G.; Antson, A.A.; Sluchanko, N.N. Molecular basis for the recognition of steroidogenic acute regulatory protein by the 14-3-3 protein family. bioRxiv 2020, 287, 3944–3966.
- Moore, B.W. Specific acidic proteins of the nervous system. In Physiological and Biochemical Aspects of Nervous Integration; Prentice-Hall: New York, NY, USA, 1967; pp. 343–359.
- Aitken, A.; Collinge, D.; Van Heusden, B.; Isobe, T.; Roseboom, P.; Rosenfeld, G.; Soll, J. 14-3-3 proteins. a highly conserved, widespread family of eukaryotic proteins. Trends Biochem. Sci. 1992, 17, 498–501.
- Wang, X.; Ren, Y.; Li, J.; Ji, Z.; Chen, F.; Wang, X. Identification of the 14-3-3 β/α-A protein as a novel maternal peptidoglycan-binding protein that protects embryos of zebrafish against bacterial infections. Dev. Comp. Immunol. 2020, 114, 103867.
- De, S. The 14-3-3 (YWHA) Proteins in mammalian reproduction. Int. Ann. Sci. 2020, 10, 52–59.
- Nathan, K.G.; Lal, S.K. The multifarious role of 14-3-3 family of proteins in viral replication. Viruses 2020, 12, 436.
- Fu, H.; Subramanian, R.R.; Masters, S.C. 14-3-3 proteins: Structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 2000, 40, 617–647.
- Pennington, K.L.; Chan, T.Y.; Torres, M.P.; Andersen, J.L. The dynamic and stress-adaptive signaling hub of 14-3-3: Emerging mechanisms of regulation and context-dependent protein-protein interactions. Oncogene 2018, 37, 5587–5604.
- Gu, Q.; Cuevas, E.; Raymick, J.; Kanungo, J.; Sarkar, S. Downregulation of 14-3-3 proteins in Alzheimer’s Disease. Mol. Neurobiol. 2020, 57, 32–40.
- Wakabayashi, K.; Umahara, T.; Hirokawa, K.; Hanyu, H.; Uchihara, T. 14-3-3 protein sigma isoform co-localizes with phosphorylated α-synuclein in Lewy bodies and Lewy neurites in patients with Lewy body disease. Neurosci. Lett. 2018, 674, 171–175.
- Wang, W.; Shakes, D.C. Molecular evolution of the 14-3-3 protein family. J. Mol. Evol. 1996, 43, 384–398.
- Rosenquist, M.; Alsterfjord, M.; Larsson, C.; Sommarin, M. Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes. Expression is demonstrated for two out of five novel genes. Plant Physiol. 2001, 127, 142–149.
- van Heusden, G.P.H. 14-3-3 proteins: Insights from genome-wide studies in yeast. Genomics 2009, 94, 287–293.
- van Heusden, G.P.; Steensma, H.Y. Yeast 14-3-3 proteins. Yeast 2006, 23, 159–171.
- Jones, D.H.; Ley, S.; Aitken, A. Isoforms of 14-3-3 protein can form homo-and heterodimers in vivo and in vitro: Implications for function as adapter proteins. FEBS Lett. 1995, 368, 55–58.
- Aitken, A.; Baxter, H.; Dubois, T.; Clokie, S.; Mackie, S.; Mitchell, K.; Peden, A.; Zemlickova, E. Specificity of 14-3-3 isoform dimer interactions and phosphorylation. Biochem. Soc. Trans. 2002, 30, 351–360.
- Gardino, A.K.; Smerdon, S.J.; Yaffe, M.B. Structural determinants of 14-3-3 binding specificities and regulation of subcellular localization of 14-3-3-ligand complexes: A comparison of the X-ray crystal structures of all human 14-3-3 isoforms. Semin. Cancer Biol. 2006, 16, 173–182.
- Chen, Y.; Ruggeri, Z.M.; Du, X. 14-3-3 proteins in platelet biology and glycoprotein Ib-IX signaling. Blood 2018, 131, 2436–2448.
- Lalle, M.; Fiorillo, A. The protein 14-3-3: A functionally versatile molecule in Giardia duodenalis. In Advances in Parasitology; Elsevier: Amsterdam, The Netherlands, 2019; Volume 106, pp. 51–103.
- Cotelle, V.; Leonhardt, N. 14-3-3 proteins in guard cell signaling. Front. Plant Sci. 2016, 6, 1210.
- Ormancey, M.; Thuleau, P.; Mazars, C.; Cotelle, V. CDPKs and 14-3-3 proteins: Emerging duo in signaling. Trends Plant Sci. 2017, 22, 263–272.
- Rittinger, K.; Budman, J.; Xu, J.; Volinia, S.; Cantley, L.C.; Smerdon, S.J.; Gamblin, S.J.; Yaffe, M.B. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell 1999, 4, 153–166.
- Sluchanko, N.N.; Gusev, N.B. Moonlighting chaperone-like activity of the universal regulatory 14-3-3 proteins. FEBS J. 2017, 284, 1279–1295.
- Tugaeva, K.V.; Kalacheva, D.I.; Cooley, R.B.; Strelkov, S.V.; Sluchanko, N.N. Concatenation of 14-3-3 with partner phosphoproteins as a tool to study their interaction. Sci. Rep. 2019, 9, 1–11.
- Yaffe, M.B.; Rittinger, K.; Volinia, S.; Caron, P.R.; Aitken, A.; Leffers, H.; Gamblin, S.J.; Smerdon, S.J.; Cantley, L.C. The structural basis for 14-3-3: Phosphopeptide binding specificity. Cell 1997, 91, 961–971.
- Coblitz, B.; Shikano, S.; Wu, M.; Gabelli, S.B.; Cockrell, L.M.; Spieker, M.; Hanyu, Y.; Fu, H.; Amzel, L.M.; Li, M. C-terminal recognition by 14-3-3 proteins for surface expression of membrane receptors. J. Biol. Chem. 2005, 280, 36263–36272.
- Trcka, F.; Durech, M.; Vankova, P.; Vandova, V.; Simoncik, O.; Kavan, D.; Vojtesek, B.; Muller, P.; Man, P. The interaction of the mitochondrial protein importer TOMM34 with HSP70 is regulated by TOMM34 phosphorylation and binding to 14-3-3 adaptors. J. Biol. Chem. 2020, 295, 8928–8944.
- Petosa, C.; Masters, S.C.; Bankston, L.A.; Pohl, J.; Wang, B.; Fu, H.; Liddington, R.C. 14-3-3ζ binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J. Biol. Chem. 1998, 273, 16305–16310.
- Masters, S.C.; Pederson, K.J.; Zhang, L.; Barbieri, J.T.; Fu, H. Interaction of 14-3-3 with a nonphosphorylated protein ligand, exoenzyme S of Pseudomonas aeruginosa. Biochemistry 1999, 38, 5216–5221.
- Henriksson, M.L.; Trollér, U.; Hallberg, B. 14-3-3 proteins are required for the inhibition of Ras by exoenzyme S. Biochem. J. 2000, 349, 697–701.
- Mils, V.; Baldin, V.; Goubin, F.; Pinta, I.; Papin, C.; Waye, M.; Eychene, A.; Ducommun, B. Specific interaction between 14-3-3 isoforms and the human CDC25B phosphatase. Oncogene 2000, 19, 1257–1265.
- Zhai, J.; Lin, H.; Shamim, M.; Schlaepfer, W.W.; Cañete-Soler, R. Identification of a novel interaction of 14-3-3 with p190RhoGEF. J. Biol. Chem. 2001, 276, 41318–41324.
- Xu, Y.; Ren, J.; He, X.; Chen, H.; Wei, T.; Feng, W. YWHA/14-3-3 proteins recognize phosphorylated TFEB by a noncanonical mode for controlling TFEB cytoplasmic localization. Autophagy 2019, 15, 1017–1030.
- Bonnefoy-Bérard, N.; Liu, Y.C.; von Willebrand, M.; Sung, A.; Elly, C.; Mustelin, T.; Yoshida, H.; Ishizaka, K.; Altman, A. Inhibition of phosphatidylinositol 3-kinase activity by association with 14-3-3 proteins in T cells. Proc. Natl. Acad. Sci. USA 1995, 92, 10142–10146.
- Garnett, M.J.; Rana, S.; Paterson, H.; Barford, D.; Marais, R. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol. Cell 2005, 20, 963–969.
- Rushworth, L.K.; Hindley, A.D.; O’Neill, E.; Kolch, W. Regulation and role of Raf-1/B-Raf heterodimerization. Mol. Cell. Biol. 2006, 26, 2262–2272.
- Ford, J.C.; Al-Khodairy, F.; Fotou, E.; Sheldrick, K.S.; Griffiths, D.; Carr, A.M. 14-3-3 protein homologs required for the DNA damage checkpoint in fission yeast. Science 1994, 265, 533–535.
- Peng, C.-Y.; Graves, P.R.; Thoma, R.S.; Wu, Z.; Shaw, A.S.; Piwnica-Worms, H. Mitotic and G2 checkpoint control: Regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 1997, 277, 1501–1505.
- Sanchez, Y.; Wong, C.; Thoma, R.S.; Richman, R.; Wu, Z.; Piwnica-Worms, H.; Elledge, S.J. Conservation of the Chk1 checkpoint pathway in mammals: Linkage of DNA damage to Cdk regulation through Cdc25. Science 1997, 277, 1497–1501.
- Bulavin, D.V.; Higashimoto, Y.; Demidenko, Z.N.; Meek, S.; Graves, P.; Phillips, C.; Zhao, H.; Moody, S.A.; Appella, E.; Piwnica-Worms, H. Dual phosphorylation controls Cdc25 phosphatases and mitotic entry. Nat. Cell Biol. 2003, 5, 545–551.
- Chen, M.-S.; Ryan, C.E.; Piwnica-Worms, H. Chk1 kinase negatively regulates mitotic function of Cdc25A phosphatase through 14-3-3 binding. Mol. Cell. Biol. 2003, 23, 7488–7497.
- Samuel, T.; Weber, H.O.; Rauch, P.; Verdoodt, B.; Eppel, J.-T.; McShea, A.; Hermeking, H.; Funk, J.O. The G2/M regulator 14-3-3ς prevents apoptosis through sequestration of Bax. J. Biol. Chem. 2001, 276, 45201–45206.
- Sunayama, J.; Tsuruta, F.; Masuyama, N.; Gotoh, Y. JNK antagonizes Akt-mediated survival signals by phosphorylating 14-3-3. J. Cell Biol. 2005, 170, 295–304.
- Zha, J.; Harada, H.; Yang, E.; Jockel, J.; Korsmeyer, S.J. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-XL. Cell 1996, 87, 619–628.
- Zhang, L.; Chen, J.; Fu, H. Suppression of apoptosis signal-regulating kinase 1-induced cell death by 14-3-3 proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 8511–8515.
- Phan, L.; Chou, P.-C.; Velazquez-Torres, G.; Samudio, I.; Parreno, K.; Huang, Y.; Tseng, C.; Vu, T.; Gully, C.; Su, C.-H. The cell cycle regulator 14-3-3σ opposes and reverses cancer metabolic reprogramming. Nat. Commun. 2015, 6, 7530.
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899.
- Ferguson, A.T.; Evron, E.; Umbricht, C.B.; Pandita, T.K.; Chan, T.A.; Hermeking, H.; Marks, J.R.; Lambers, A.R.; Futreal, P.A.; Stampfer, M.R. High frequency of hypermethylation at the 14-3-3 σ locus leads to gene silencing in breast cancer. Proc. Natl. Acad. Sci. USA 2000, 97, 6049–6054.
- Lee, M.-H.; Lozano, G. Regulation of the p53-MDM2 pathway by 14-3-3 σ and other proteins. Semin. Cancer Biol. 2006, 16, 225–234.
- Yang, H.; Zhao, R.; Lee, M.-H. 14-3-3σ, a p53 regulator, suppresses tumor growth of nasopharyngeal carcinoma. Mol. Cancer Ther. 2006, 5, 253–260.
- Yang, H.-Y.; Wen, Y.-Y.; Chen, C.-H.; Lozano, G.; Lee, M.-H. 14-3-3σ positively regulates p53 and suppresses tumor growth. Mol. Cell. Biol. 2003, 23, 7096–7107.
- West-Foyle, H.; Kothari, P.; Osborne, J.; Robinson, D.N. 14-3-3 proteins tune non-muscle myosin II assembly. J. Biol. Chem. 2018, 293, 6751–6761.
- Chan, T.A.; Hermeking, H.; Lengauer, C.; Kinzler, K.W.; Vogelstein, B. 14-3-3σ is required to prevent mitotic catastrophe after DNA damage. Nature 1999, 401, 616–620.
- Laronga, C.; Yang, H.-Y.; Neal, C.; Lee, M.-H. Association of the cyclin-dependent kinases and 14-3-3 sigma negatively regulates cell cycle progression. J. Biol. Chem. 2000, 275, 23106–23112.
- Hong, H.-Y.; Jeon, W.-K.; Bae, E.-J.; Kim, S.-T.; Lee, H.-J.; Kim, S.-J.; Kim, B.-C. 14-3-3 sigma and 14-3-3 zeta plays an opposite role in cell growth inhibition mediated by transforming growth factor-beta 1. Mol. Cells 2010, 29, 305–309.
- Hong, H.-Y.; Jeon, W.-K.; Kim, S.-J.; Kim, B.-C. 14-3-3 σ is a new target up-regulated by transforming growth factor-β1 through a Smad3-dependent mechanism. Biochem. Biophys. Res. Commun. 2013, 432, 193–197.
- Ingles-Esteve, J.; Morales, M.; Dalmases, A.; Garcia-Carbonell, R.; Jene-Sanz, A.; López-Bigas, N.; Iglesias, M.; Ruiz-Herguido, C.; Rovira, A.; Rojo, F. Inhibition of specific NF-κB activity contributes to the tumor suppressor function of 14-3-3σ in breast cancer. PLoS ONE 2012, 7, e38347.
- Wolter, M.; de Vink, P.; Neves, J.F.; Srdanovic, S.; Higuchi, Y.; Kato, N.; Wilson, A.J.; Landrieu, I.; Brunsveld, L.; Ottmann, C. Selectivity via cooperativity: Preferential stabilization of the p65/14-3-3 interaction with semi-synthetic natural products. J. Am. Chem. Soc. 2020, 142, 11772–11783.
- Aghazadeh, Y.; Papadopoulos, V. The role of the 14-3-3 protein family in health, disease, and drug development. Drug Discov. Today 2016, 21, 278–287.
- Anders, C.; Higuchi, Y.; Koschinsky, K.; Bartel, M.; Schumacher, B.; Thiel, P.; Nitta, H.; Preisig-Müller, R.; Schlichthörl, G.; Renigunta, V.; et al. A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface. Chem. Biol. 2013, 20, 583–593.
- Zúñiga, R.; Valenzuela, C.; Concha, G.; Brown, N.; Zúñiga, L. TASK-3 downregulation triggers cellular senescence and growth inhibition in breast cancer cell lines. Int. J. Mol. Sci. 2018, 19, 1033.
- Guillory, X.; Wolter, M.; Leysen, S.; Neves, J.F.; Kuusk, A.; Genet, S.; Somsen, B.; Morrow, J.; Rivers, E.; van Beek, L. Fragment-based differential targeting of PPI stabilizer interfaces. J. Med. Chem. 2020, 63, 6694–6707.
- Zhou, X.; Lei, Q.-Y. Regulation of TAZ in cancer. Protein Cell 2016, 7, 548–561.
- Lodygin, D.; Yazdi, A.; Sander, C.; Herzinger, T.; Hermeking, H. Analysis of 14-3-3 expression in hyperproliferative skin diseases reveals selective loss associated with CpG-methylation in basal cell carcinoma. Oncogene 2003, 22, 5519–5524.
- Li, Z.; Liu, J.-Y.; Zhang, J.-T. 14-3-3σ, the double-edged sword of human cancers. Am. J. Transl. Res. 2009, 1, 326.
- Chan, S.Y.-Y.; To, K.-F.; Leung, S.-F.; Yip, W.W.-L.; Mak, M.K.-F.; Chung, G.T.-Y.; Lo, K.-W. 14-3-3σ expression as a prognostic marker in undifferentiated nasopharyngeal carcinoma. Oncol. Rep. 2010, 24, 949–955.
- Qi, Y.-J.; Wang, M.; Liu, R.-M.; Wei, H.; Chao, W.-X.; Zhang, T.; Lou, Q.; Li, X.-M.; Ma, J.; Zhu, H. Downregulation of 14-3-3σ correlates with multistage carcinogenesis and poor prognosis of esophageal squamous cell carcinoma. PLoS ONE 2014, 9, e95386.
- Zhou, R.; Shao, Z.; Liu, J.; Zhan, W.; Gao, Q.; Pan, Z.; Wu, L.; Xu, L.; Ding, Y.; Zhao, L. COPS5 and LASP1 synergistically interact to downregulate 14-3-3σ expression and promote colorectal cancer progression via activating PI3K/AKT pathway. Int. J. Cancer 2018, 142, 1853–1864.
- Umbricht, C.B.; Evron, E.; Gabrielson, E.; Ferguson, A.; Marks, J.; Sukumar, S. Hypermethylation of 14-3-3 σ (stratifin) is an early event in breast cancer. Oncogene 2001, 20, 3348–3353.
- Vercoutter-Edouart, A.-S.; Lemoine, J.; Le Bourhis, X.; Louis, H.; Boilly, B.; Nurcombe, V.; Révillion, F.; Peyrat, J.-P.; Hondermarck, H. Proteomic analysis reveals that 14-3-3σ is down-regulated in human breast cancer cells. Cancer Res. 2001, 61, 76–80.
- Urano, T.; Saito, T.; Tsukui, T.; Fujita, M.; Hosoi, T.; Muramatsu, M.; Ouchi, Y.; Inoue, S. Efp targets 14-3-3σ for proteolysis and promotes breast tumour growth. Nature 2002, 417, 871–875.
- Choi, H.H.; Gully, C.; Su, C.-H.; Velazquez-Torres, G.; Chou, P.-C.; Tseng, C.; Zhao, R.; Phan, L.; Shaiken, T.; Chen, J. COP9 signalosome subunit 6 stabilizes COP1, which functions as an E3 ubiquitin ligase for 14-3-3σ. Oncogene 2011, 30, 4791.
- Uchida, D.; Begum, N.; Almofti, A.; Kawamata, H.; Yoshida, H.; Sato, M. Frequent downregulation of 14-3-3 σ protein and hypermethylation of 14-3-3 σ gene in salivary gland adenoid cystic carcinoma. Br. J. Cancer 2004, 91, 1131–1138.
- Yi, B.; Tan, S.X.; Tang, C.E.; Huang, W.G.; Cheng, A.L.; Li, C.; Zhang, P.F.; Li, M.Y.; Li, J.L.; Yi, H. Inactivation of 14-3-3 σ by promoter methylation correlates with metastasis in nasopharyngeal carcinoma. J. Cell. Biochem. 2009, 106, 858–866.
- Sun, L.; Ain, Q.U.; Gao, Y.-S.; Khan, G.J.; Yuan, S.-t.; Roy, D. Effect of Marsdenia tenacissima extract on G2/M cell cycle arrest by upregulating 14-3-3σ and downregulating c-myc in vitro and in vivo. Chin. Herb. Med. 2019, 11, 169–176.
- Han, B.; Xie, H.; Chen, Q.; Zhang, J.-T. Sensitizing hormone-refractory prostate cancer cells to drug treatment by targeting 14-3-3σ. Mol. Cancer Ther. 2006, 5, 903–912.
- Liu, Y.; Liu, H.; Han, B.; Zhang, J.-T. Identification of 14-3-3σ as a contributor to drug resistance in human breast cancer cells using functional proteomic analysis. Cancer Res. 2006, 66, 3248–3255.
- Zhang, J.-T.; Liu, Y. Use of comparative proteomics to identify potential resistance mechanisms in cancer treatment. Cancer Treat. Rev. 2007, 33, 741–756.
- Ghahary, A.; Marcoux, Y.; Karimi-Busheri, F.; Li, Y.; Tredget, E.E.; Kilani, R.T.; Lam, E.; Weinfeld, M. Differentiated keratinocyte-releasable stratifin (14-3-3 sigma) stimulates MMP-1 expression in dermal fibroblasts. J. Investig. Dermatol. 2005, 124, 170–177.
- Thiel, P.; Roglin, L.; Meissner, N.; Hennig, S.; Kohlbacher, O.; Ottmann, C. Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions. Chem. Commun. (Camb.) 2013, 49, 8468–8470.
- Corradi, V.; Mancini, M.; Santucci, M.; Carlomagno, T.; Sanfelice, D.; Mori, M.; Vignaroli, G.; Falchi, F.; Manetti, F.; Radi, M.; et al. Computational techniques are valuable tools for the discovery of protein-protein interaction inhibitors: The 14-3-3σ case. Bioorg. Med. Chem. Lett. 2011, 21, 6867–6871.
- Iralde-Lorente, L.; Tassone, G.; Clementi, L.; Franci, L.; Munier, C.C.; Cau, Y.; Mori, M.; Chiariello, M.; Angelucci, A.; Perry, M.W. Identification of phosphate-containing compounds as new inhibitors of 14-3-3/c-Abl protein-protein interaction. ACS Chem. Biol. 2020, 15, 1026–1035.
- Iralde-Lorente, L.; Tassone, G.; Clementi, L.; Franci, L.; Munier, C.C.; Cau, Y.; Mori, M.; Chiariello, M.; Angelucci, A.; Perry, M.W. Identification of phosphate-containing compounds as new inhibitors of 14-3-3/c-Abl protein-protein interaction. ACS Chem. Biol. 2020, 15, 1026–1035.




