Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Alexandra Elisabeta Avanu | -- | 4252 | 2024-03-19 18:07:45 | | | |
| 2 | Fanny Huang | -1 word(s) | 4251 | 2024-03-20 04:49:44 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Avanu, A.E.; Ciubotariu, A.M.; Dodi, G. Oligonucleotide-Based Biosensors in Early Diagnosis of Gastric Cancer. Encyclopedia. Available online: https://encyclopedia.pub/entry/56410 (accessed on 08 February 2026).
Avanu AE, Ciubotariu AM, Dodi G. Oligonucleotide-Based Biosensors in Early Diagnosis of Gastric Cancer. Encyclopedia. Available at: https://encyclopedia.pub/entry/56410. Accessed February 08, 2026.
Avanu, Alexandra E., Alexandra M. Ciubotariu, Gianina Dodi. "Oligonucleotide-Based Biosensors in Early Diagnosis of Gastric Cancer" Encyclopedia, https://encyclopedia.pub/entry/56410 (accessed February 08, 2026).
Avanu, A.E., Ciubotariu, A.M., & Dodi, G. (2024, March 19). Oligonucleotide-Based Biosensors in Early Diagnosis of Gastric Cancer. In Encyclopedia. https://encyclopedia.pub/entry/56410
Avanu, Alexandra E., et al. "Oligonucleotide-Based Biosensors in Early Diagnosis of Gastric Cancer." Encyclopedia. Web. 19 March, 2024.
Copy Citation
Gastric cancer (GC) remains a significant global health challenge, with late-stage diagnosis impacting treatment options and decreased survival rates. To address this, there has been a growing interest in the development of innovative screening and diagnostic methods. Nanobiosensors have undergone multiple iterations and unveiled remarkable features that pledge to reshape patient care.
biosensors
nanotechnology
DNA
RNA
early cancer detection
gastric cancer
cancer diagnosis
nano
1. Introduction
According to the Globocan database, gastric cancer (GC) stands as a global health problem, with around 1 million new cases diagnosed annually, resulting in almost 700,000 deaths in 2022 and ranking fifth worldwide in terms of mortality rates in both sexes and all ages [1]. The regions most affected are Asia, with a staggering percentage of over 70% and Europe, with around 14.5%, in both incidence and fatalities [1][2][3].
The aggressive nature of GC, coupled with its poor prognosis, underscores the critical importance of early detection for effective intervention [1]. It is crucial to highlight that with early-stage treatment, survival rates can reach an impressive 92.6% [4][5]. In Japan, early GC (EGC) diagnosis achieves 50%, yielding a 90% 5-year survival rate [6]. On the flip side, for cases at an advanced stage, it ranges from only 10 to 30% [4][5] and it is associated, more often than not, with severe complications [4]. Global mortality rates have shown minor decreases in the last 4 decades [7][8]. For example, in regions like North America and Latin American countries, mortality rates decreased by around 2% annually, between 1980 and 2011. In Europe, the decline was at 3% in the same time span. Because of the lower global incidence, GC screening (by X-rays, endoscopies, etc.) only takes place in affluent Asian countries, more specifically, in Japan and Korea. However, the global burden of GC is substantial, with a 2018 study, across 31 European countries, estimating its costs at around EUR 5 billion, originating from healthcare spending and productivity losses [9][10], emphasizing the need for informed decisions to enhance cancer care.
Traditional and current diagnostic practices are upper gastrointestinal (UGI) endoscopy followed by biopsy sampling and histopathological examination [2][11], with 9.4% of cancers missed, as demonstrated by a meta-analysis by Pimenta-Melo et al. (2016) involving over 69,000 patients [12][13]. Some of the reasons are a detectable tumor size requirement of ≥5 mm [14], younger age, gender and the gastric pathologies trio, atrophy, adenoma or ulcer [12][13]. Recent advancements in biomedical sciences have led to the development of many tumor marker determination methods. Immunoassay techniques, such as radioimmunoassay (RIA) and enzyme-linked immunosorbent assay (ELISA), became cornerstones in clinical quantitative tumor marker detection [15]. Despite their prevalence, supported by 95% to 99% accuracy [2][16], these methods are hindered by time-consuming processes, high cost and a dependency on sophisticated instrumentation and skilled personnel [11][17]. Furthermore, protein biomarkers detected in such tests often lack adequate specificity [4] and sensitivity [18].
To increase reliability, it was proposed that multiple biomarkers be used [6]. For instance, Wang et al. (2022) considered the most frequent tumor protein biomarkers used in the clinic, the carcinoembryonic antigen (CEA), the carbohydrate antigen 19-9 (CA19-9) and the tumor-associated glycoprotein 72-4 (CA72-4), and showed a combined specificity of 89% and a sensitivity of 67% [18], while still requiring ample resources for analysis [6].
Another significant focus is on Helicobacter pylori (H. pylori), a well-known carcinogen. Studies showed that its eradication reduces the risk of GC [19]. The issues yet to be addressed are the accurate identification of screening and eradication target populations and antibiotic overuse and resistance, among others. Other biomarkers in use are serum pepsinogens (PGs), which unfortunately identify only gastric precancerous lesions and not GC [6], death-ligand 1 (PD-L1) with only 62% sensitivity and 73% specificity [20] and low prevalence cluster of differentiation 44 variant 9 (CD44-v9), which does not serve as a prognostic biomarker in advanced GC, except in the early stages [21].
In the quest of surmounting these impediments, the preclinical sector turned to nanotechnology, a field with significant breakthroughs in the past decade, allowing the integration of diverse diagnostic modalities into a unified platform. There is evidence that sensitivity could increase 20 times more when compared to ELISA [22][23]. On one hand, in conjunction with a variety of analytical methods, it has propelled the creation of multifunctional nanocarriers-based biological sensors and on the other hand there are chemical sensors, or chemosensors for short and together they have the collective goal of optimizing their performance for low-cost, portable, on-site clinical setting diagnosis [24]. Their superior effectiveness is attributed to the specific combination of analytes, bioactive substances (biomarkers or biocomponents) and transducers, converting biological responses into electrical signals [11]. They exhibit compelling viability that is further fortified by accessible samples or analytes, such as patient’s saliva, urine, serum, plasma and more, as shown in Figure 1, thereby expediting real-time monitoring capabilities. This has given the cancer research community a beacon of hope for improved diagnostic accuracy and timely intervention.
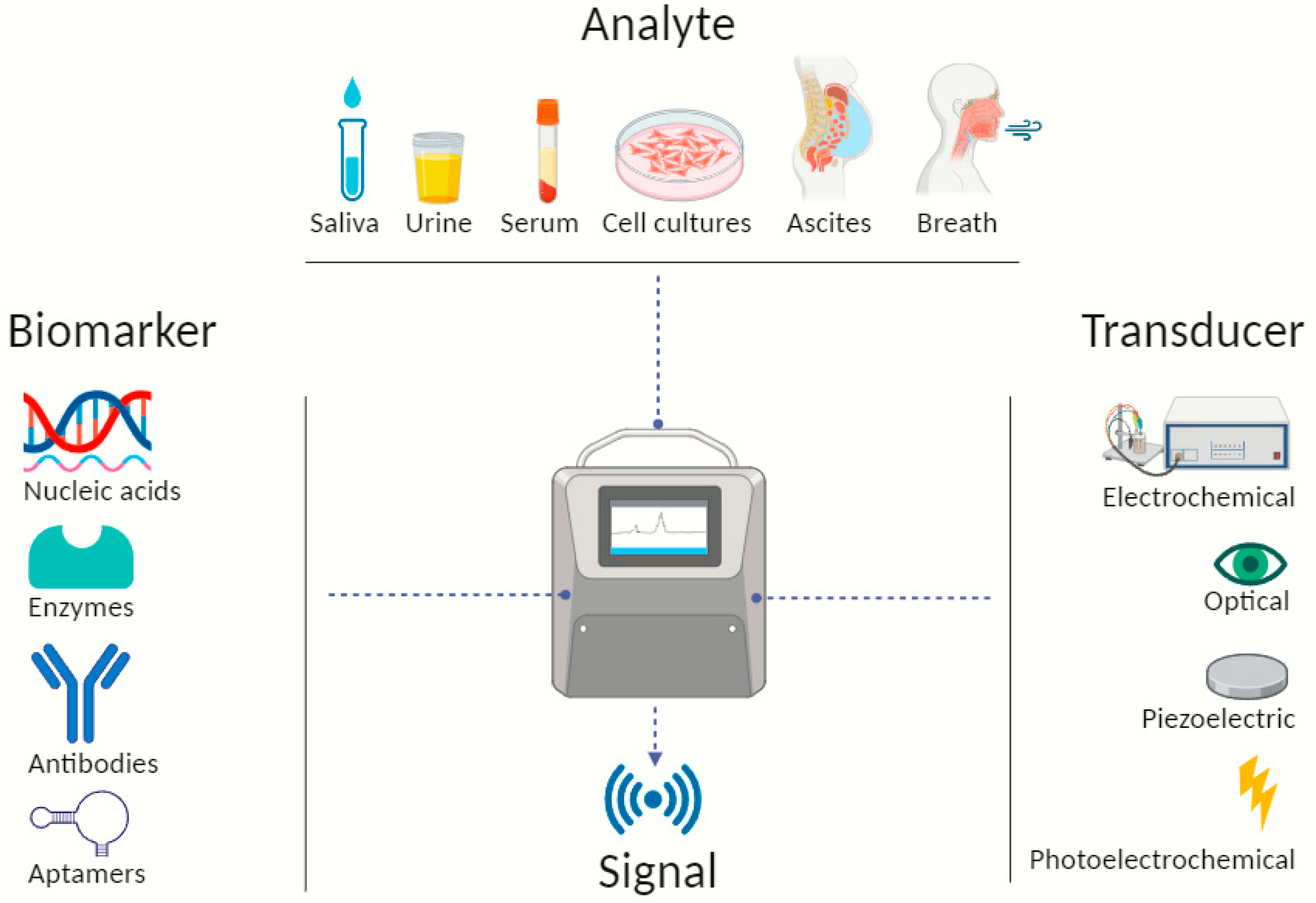
Figure 1. Illustrative scheme describing typical biosensor components detecting and identifying biomarkers from different types of analytes. Created with BioRender.com.
The development of portable and miniaturized devices suitable for point-of-care applications that allow on-site testing, reducing the need for centralized laboratories and enabling faster decision-making in medical or field settings, in an affordable fashion, is therefore required.
Although nanotechnology has not yet been deployed clinically for cancer diagnosis, it is already on the market in a variety of medical tests and screens, including home pregnancy tests that use gold nanoparticles [25], blood sugar and cholesterol level tracking, infectious disease identification and other additional applications [26].
2. DNA Nanobiosensors in Gastric Cancer
Biosensors can be engineered to rapidly recognize and bind specifically to target genes, mutations or pathogenic sequences in real-time [27]. This specificity is achieved through the design of molecular probes or recognition elements that are complementary to the target DNA. This ensures that the biosensor responds only to the presence of the desired DNA sequence, eliminating cross-reactivity. Additionally, many DNA biosensors exhibit high sensitivity, allowing for detection at low concentrations. This is crucial for applications such as early disease diagnosis, where only trace amount of DNA may be present.
DNA biosensors are often compatible with various detection platforms, including optical, electrochemical, and microfluidic systems [28]. This versatility allows for the integration of DNA biosensors into different analytical devices and technologies.
The usual need for amplification techniques such as polymerase chain reaction (PCR), which uses thermal cycling and loop-mediated isothermal amplification (LAMP) that has the requirement of a relatively complex primer design [29], has been replaced with easier and more sensitive techniques leading to lower detection limits.
In Table 1, the features of nanobiosensors that detect DNA in human GC samples are summarized. Researchers covered details like the operating principle, transducers and their associations with materials like gold nanoparticles, carbon nanotubes and quantum dots. It is important to emphasize that all nanosensors from this table showed rapid response times, high sensitivity, successful optimization and good economical attributes.
Table 1. DNA nanobiosensors for GC.
| Sensing Platform | Transducer | Biomarker | Human Sample | LoD | Takeaways | Ref. |
|---|---|---|---|---|---|---|
| High-density “hot spot” AuNPs@SiO array substrate with RCA strategy | Optical (SERS) | M.SssI | Serum | 2.51 × 10−4 U mL−1 | Simple preparation, high biocompatibility, uniformity, reproducibility, stability | [30] |
| Polymeric l-arginine and rGO-AuNSs on glass electrode | Electrochemical (CV) | PIK3CA ctDNA | 1.0 × 10−20 M | Label-free, desirable stability, wide dynamic response | [31] | |
| SWCN DMEJ with DNA–gold urchin | Electrochemical (IDE) | SOX-17 | 1 aM | High performance, efficiency, biocompatibility, no cross-reactivity | [32] | |
| Nitrophenyl-functionalized black phosphorus nanosheets and FAM labelling | Optical (fluorescence) | PIK3CA E542K ctDNA | Tumor cell lines | 50 fM | Enzyme-free, long-term stability, simple manufacturing process, good discrimination ability of interferences | [33] |
| Nanoplasmonic, nanogold-linked sorbent assay | Optical (FOPPR and FONLISA) | Methylated SOCS-1 | Tumor tissue and cell lines | 0.81 fM | PCR- and amplification-free, label- and sequencing-free; superior to PCR and other assays | [34] |
Abbreviations: aM—attomolar; GC—gastric cancer; LoD—limit of detection; Ref—references; AuNPs@SiO2—gold silica nanoparticles; RCA—rolling circle amplification; SERS—surface-enhanced Raman scatting; M.SssI—CpG methyltransferase; rGO-AuNSs—graphene oxide-wrapped gold nanostars; CV—cyclic voltammetry; ctDNA—circulating tumor DNA; SWCN—single-walled carbon nanotube; DMEJ—different dimicroelectrodes junction; IDE—interdigitated electrode; FAM—carboxyfluorescein; fM—femtomolar; FOPPR—fiber optic particle plasmon resonance; FONLISA—fiber optic nanogold-linked immunosorbent assay; PCR—polymerase chain reaction.
According to several studies, irregular changes in the DNA methylation pattern, which normally regulates gene activity and cell differentiation, can serve as a valuable biomarker for EGC detection [35][36]. Hypermethylation contributes to GC and cancers in general, by silencing tumor suppressor genes (TSGs) [37]. Guthula et al. (2022) effectively tackled numerous challenges listed in Table 1 associated with detecting DNA methylation in the frequently inactivated SOCS-1 human genome, a gene linked to cancers such as GC [34]. They created a rapid (≤15 min) PCR-free sensor that exhibited a strong correlation with PCR outcomes and the lowest LoD among amplification-free methods reported previously, affirming the reliability of their approach. Ge et al. (2021) even went a step further back, looking into CpG methyltransferase targeting, which accumulates before proceeding to participate in DNA methylation [30]. Boasting high accuracy, selectivity, and sensitivity, the authors engineered a biosensor that had a particularly low LoD. They employed surface-enhanced Raman scattering (SERS) as the transduction method, surpassing traditional approaches that demand substantial sample quantities. SERS biosensors are developed on the enhancement of Raman scattering signals that occur when molecules are in close proximity to specially designed surfaces featuring nanoscale metallic structures, such as gold or silver nanoparticles [38]. Besides the known addressed limitations of SERS related to substrate preparation, uniformity, external factors and signal fluctuations, the shelf-life question remains unanswered [39]. Also, some argue that it can be laborious and not easily portable [40].
To improve the detection of target DNA, in 2021, Yu et al. attached a DNA sequence from the SOX-17 gene onto a gold urchin (DNA-GU), linking it to a single-walled carbon nanotube (SWCN)-constructed DMEs junction (DMEJ) [32]. The biosensor demonstrated exceptional sensitivity, detecting DNA concentrations ranging from 1 aM to 10 fM. The successful detection was attributed to the strategic immobilization of the capture molecule, aligning with research showing that higher-density biomolecules enhance sensor performance. Showcasing its selective identification capabilities, the biosensor effectively differentiated target DNA from complementary sequences, including miR-106a, the subject of numerous RNA nanosensors.
Introducing another significant biomarker, the circulating tumor DNA (ctDNA) refers to a class of circulating free DNA shed by the tumor cells, desired to provide insights into gastric tumor presence and dynamics through liquid biopsy sampling [41]. Liquid biopsies involve the analysis of blood or other body fluids, eliminating the need for invasive procedures like traditional tissue biopsies [42]. This makes them more suitable for point-of-care testing, which prioritizes quick and minimally invasive diagnostic methods. Rahman et al. (2022) studied the precise diagnosis of GC through hybridization between the capturing DNA probe and PIK3CA gene of ctDNA specimens obtained via liquid biopsy [31]. They chose to combine graphene oxide with a large surface area and star-like shaped gold nanostructures (AuNSs). The material was deposited onto a glass electrode, forming a thin layer of coating, and the outcome showed great promise.
For the same biomarker, Huang et al. (2020) developed a biosensor that leveraged nitrophenyl-functionalized black phosphorus nanosheets (NP-BPs) [33]. It was constructed on the foundation of surface-modified BPs, discriminated well between different DNA structures. Practical experiments revealed exceptional sensitivity, detecting ctDNA concentrations as low as 50 fM, with a broad linear detection range of 50 fM to 80 picomolar (pM). The biosensor’s clinical application was demonstrated by successfully detecting ctDNA in clinical serum samples, presenting a LoD of 0.5 nanomolar. Furthermore, the biosensor’s performance was enhanced by combining it with conventional magnetic extraction, achieving a lower detection limit of 50 fM. While the assay offered advantages like a 15 min speed and simplicity, its sensitivity fell short compared to that of PCR.
The previous study and others have explored different methods for synthesizing nanocomposites, such as graphene/metal oxide, graphite electrodes plus various metals and metal oxides. These cannot be applied directly in detecting ctDNA in serum samples due to the interference from the strong nonspecific absorption of serum proteins. In this context, Ma et al. (2020) presented a one-step strategy for preparing zinc-based nanohybrids with tunable structures [43]. The proposed approach involved the use of conducting polypyrrole (PPy) as a heating source under microwave irradiation for PIK3CA gene detection. Additionally, the nanocomposites showed a reliable performance in distinguishing mismatches in DNA, highlighting their applicability in detecting genetic variations associated with GC.
Cao et al. (2022) present another notable example, albeit in the preclinical stage and conducted on mice serum, proposing the detection of the PIK3CA gene in ctDNA [40]. They created a microfluidic chip for SERS, pursuing PIK3CA E542K and TP53 genes detection. The removal of enzymes as catalysts which are usually used in amplification strategies like RCA led to the combination of two enzyme-free signal amplification strategies, namely the catalytic hairpin assembly (CHA) and hybridization chain reaction (HCR) in order to overcome insufficient signal gain and sensitivity [40]. The study conducted stands out in the realm of microfluidic methods, achieving an exceptionally favorable LoD in the aM range, with reported values of 1.26 aM and 2.04 aM, respectively, and a detection speed of 13 min.
However, in 2022, Dang et al. affirmed that ctDNA’s practical value in the clinical setting is yet to be established [44]. Its absence, emphasize the authors, cannot definitively rule out GC or other types of cancer. In 2023, Bittla et al. (2023) also sternly concluded in a systematic review that despite expectations and efforts, ctDNA has not demonstrated its usefulness in cancer detection but could be used in the future only as a predictor [45].
These diverse biosensing approaches demonstrate both progress and challenges in the quest for effective and reliable diagnostic tools for GC. Future research should focus on addressing remaining challenges, such as shelf life or limitations of specific nanomaterials.
3. RNA Nanobiosensors in Gastric Cancer
Alterations to typical characteristics of normal cells, such as to microRNAs (miRs), are considered RNA-based cancer biomarkers [46]. Although critical to cell physiology, miRs, small non-coding RNAs, act as molecular signatures for cancer detection and are linked to cancer stage, tumor size and cell proliferation. These molecules can persist in a detectable and consistent manner, making them reliable biomarkers [47]. Various methods, including electrochemical methods, optical methods or the combination of the two, using nanotechnology have been explored for GC detection. Conventional techniques like PCR or Northern blot, while capable of identifying RNA biomarkers, have limitations and lack sensitivity [48]. Table 2 provides an overview of the most recent nanobiosensors detecting RNA, highlighting their key features in EGC diagnosis. The same principle of adding only distinct and supplementary characteristics was again consistently applied. Having said that, each sensor exhibited swift detection times, elevated sensitivity and appreciable cost-effectiveness.
Table 2. RNA nanobiosensors in GC.
| Sensing Platform | Transducer | Biomarker | Human Sample | LoD | Takeaways | Ref. |
|---|---|---|---|---|---|---|
| Blackberry-like magnetic DNA/FMMA nanospheres on gold stir-bar using CHA-HCR and RAFT amplification | Electrochemical (V) | miR-106a | Serum | 0.68 aM | Enzyme-free, simple nanomaterials, acceptable storage stability, RNA extraction-free, sample pretreatment-free technique, high recovery | [49] |
| Gold–magnetic NPs single-strand (ss) probe 1 (P1) | Electrochemical (EIS, CV, DPV) | miR-106a | Serum | 0.3 fM | Great performance, stability, simplicity, reproducibility, agreeable storage stability | [50] |
| AuNPs and CdSe@CdS QDs-contained magnetic nanocomposites labels with polythiophene/rGO-modified carbon electrodes | Electrochemical (CV, DPV) | miR-106a let-7a |
Plasma | 0.06 fM (miR-106a) 0.02 fM (let-7a) |
Multiplexing, good recovery, reproducibility, appropriate storage stability | [51] |
| AgNRs array coated by the mF-MoS2 NSs, dual mode detection assay | Optical (SERS) and electrochemical (SWV) | miR-106a | Serum | 67.44 fM 248.01 fM |
In situ, stability, reliability, reproducibility, minimal interference | [52] |
| Perovskite–graphene oxide nanocomposite on an electrode, genosensing assay | Electrochemical (chronoamperometry) | miR-21 | Cell lines | 2.94 fM | Label-free, reproducibility, reusability, stability, versatility, robustness | [53] |
| Ratiometric strategy using CDs with triple function and FAM-labeled ssDNA | Optical (fluorescence) | miR-21 | Plasma | 1 pM | Reproducibility, reliability, simplicity, strong anti-interference ability, excellent performance | [54] |
| Two-stage cyclic enzymatic amplification with T4 RNA ligase 2 and T7 exonuclease and AuNPs | Electrochemical (DPV) | miR-21 | Serum | 0.36 fM | Convenience, reproducibility, excellent performance, stability | [55] |
| MXene-derivative QDs (Mo2TiC2 QDs) and SnS2 nanosheets/lipid bilayer | Electrochemical and optical (voltammetry and fluorescence) | miR-27a-3p | Ascites | 1 fM | Reproducibility, low background noise, wide dynamic range, good stability, minimal interference | [56] |
| “Hot spot” bismuth nano-nest/Ti3CN QD- SPC-ECL | Electrochemical and optical (voltammetry and fluorescence) | miR-421 | Ascites | 0.3 fM | Improved luminescence and catalytic activity, stability, controllability | [57] |
| Dual-response–single-amplification nanomachine | Optical (fluorescence) | miR-5585-5p & PLS3 mRNA | Serum | 1.19 fM (miR-5585-5p) 16.37 fM (PLS3) |
Enzyme-free, extraction-free, high recovery, great performance | [58] |
| CPs/AuNP-AuE with DSN | Electrochemical (chronoamperometry and CV) | miR-100 | Serum | 100 aM | Enzyme-free, reliability, controllability, effectiveness | [59] |
Abbreviations: FMMA—ferrocenylmethyl methacrylate; CHA-HCR—catalyzed hairpin assembly—hybridization chain reaction; RAFT—reversible addition fragmentation transfer; V—voltammetry; miR—microRNA; NPs—nanoparticles; EIS—electrochemical impedance spectroscopy; CV—cyclic voltammetry; DPV—differential pulse voltammetry; AuNPs—gold nanoparticles; QDs—quantum dots; rGO—reduced graphene oxide; AgNRs—Ag nanorods; mF-MoS2—multi-functionalized molybdenum disulfide nanosheet; NSs—nanostars; SERS—surface enhanced Raman scattering; SWV—square wave voltammetry; CDs—carbon dots; FAM—carboxyfluorescein; ssDNA—single-stranded DNA; Mo2TiC2—molybdenum titanium carbide; SnS2—tin sulfide; Ti3CN—titanium carbonitride; SPC-ECL—surface plasmon coupling electrochemiluminescence; CPs—capture probes; AuE—Au electrode; DSN—duplex-specific nuclease.
MiR-106a, a member of the miR-17 family, recognized as an oncogene in GC cells, exhibits a direct association with the occurrence of tumor metastasis [60]. This molecular behavior, coupled with its detectability in liquid biopsies, positions miR-106a as a compelling biomarker for biosensors. In 2016, Daneshpour et al. pioneered a nanobiosensor featuring double-probe sandwich architecture that incorporates gold–magnetic NPs [50]. This sensor demonstrated exceptional precision, sensitivity and selectivity in detecting miR-106a, showcasing prolonged stability for over 7 weeks. Building on this success, in 2018, Daneshpour et al. introduced a novel biosensing technology with multiplexing capabilities for the simultaneous detection of miR-106a and let-7a, both associated with GC [51]. The advanced multiplexed biosensing platform utilized a modified screen-printed carbon electrode (SPCE) with polythiophene (PTh), a conducting polymer and reduced graphene oxide (rGO). The procedure occurred at room temperature in physiological pH conditions. The method’s sensitivity was evaluated, revealing a low detection limit of 0.06 fM for miR-106a and 0.02 fM for let-7a. The combination of PTh and rGO layers on the SPCE surface aimed to enhance the conductivity and stability of the electrode, which was vital for improving the performance of the biosensing platform. In the same year, Park et al., introduced on-chip colorimetric biosensing for the early detection of the same biomarker [61]. The platform was based on the plasmon coupling of hybridized AuNPs showing high specificity and sensitivity. Two years after, Shafiee et al. (2020) leveraged the unique properties of DNA, such as molecular programmability and nanoscale controllability, which led to the creation of a complementary DNA strand for miR-106a [62]. DNA, renowned for its organic ligand characteristics, proved to be an excellent choice, being widely acknowledged as a fundamental building block for novel nanomaterials [63]. In both cases, despite the improved capabilities, this analysis seemed to necessitate more time than the optimal duration expected for an ideal biosensor. Nevertheless, Radfar et al. addressed this challenge in 2022 by employing a combination of CHA-HCR and RAFT polymerization for signal amplification [49]. This innovative approach significantly enhanced the sensitivity of miR-106a detection and obtained a LoD of 0.68 aM. Importantly, the authors successfully tackled the issue of shelf-life, with 94.3% of the oxidation peak current being retained after 6 weeks. Through an innovative dual transducing mode, Zhai et al. (2022) used multi-functionalized molybdenum disulfide nanosheet (mF-MoS2 NS) probes and SERS-active Ag nanorods (AgNRs) array electrode, to build an miR-106-detecting biosensor with superior reproducibility and higher sensitivity [52]. Limitations, such as using different instruments for the synchronous multimodal analysis, were successfully addressed. Samples were obtained via liquid biopsies.
Upregulated across various cancers, miR-21 acts as an oncogenic microRNA influencing multiple TSGs, and given its frequent upregulation in GC, it could serve as a potential diagnostic biomarker for GC [64]. In 2016, Li et al. employed T4 RNA ligase 2, an enzyme proficient in catalyzing the ligation of both inter- and intramolecular RNA molecules [55]. This enzyme was utilized to initiate a specific ligation reaction based on the target RNA sequence. Additionally, T7 exonuclease, known for degrading single-stranded DNA in a 5′ to 3′ direction, was employed to instigate and propel the cyclic amplification of the target RNA. Through this two-stage cyclic enzymatic amplification method (CEAM), the researchers successfully detected miRNA-21 at a low concentration of 0.36 fM, showcasing exceptional specificity. Notably, the introduction of mismatched non-complementary RNAs did not induce noticeable signal changes, affirming the success of this nanosensor. Similarly, in the biosensor designed by Wang et al. (2020) the ratiometric fluorescence strategy, along with T7 exonuclease-mediated cyclic enzymatic amplification, was employed to enhance the precision and accuracy of the detection process [54]. The use of carbon dots (CDs) and 6-carboxyfluorescein (FAM) as labels contributed to the ratiometric fluorescence approach. The results demonstrated good correlation with quantitative reverse transcription polymerase chain reaction (qRT-PCR), and notably, in healthy patients, the expression of miR-21 was significantly lower.
Another crucial oncogenic miRNA in GC was put through tests by Li et al. [56]. They utilized molybdenum titanium carbide quantum dots (Mo2TiC2 QDs) in an electrochemiluminescence (ECL) biosensor to detect GC marker miR-27a-3p. The biosensor incorporated SnS2 nanosheets and a lipid bilayer, enhancing QD luminosity and stability. The synergistic system achieved a wide miRNA-27a-3p detection range (1 fM to 10 nM) with a LoD at 1 fM.
Li et al. (2023) introduced a novel surface plasmon-coupled electrochemiluminescence (SPC-ECL) biosensor, combining Ti3CN QDs with enhanced luminescence and a specially designed bismuth nano-nest structure with strong localized surface plasmon resonance (LSPR) effects [57]. The biosensor successfully quantified miRNA-421 in a concentration range of 1 fM to 10 nM and demonstrated clinical applicability using ascites samples from GC patients. Of course, minimally invasive or non-invasive sampling approaches are preferred.
Multiplexing is pivotal for biosensors as it enables the simultaneous detection of multiple analytes, enhancing efficiency and providing comprehensive diagnostic information [65]. Zhang et al. (2023) introduced an innovative dual-target responsive fluorescent nanomachine for the simultaneous detection of miR-5585-5p and PLS3 mRNA [58]. Guided by advanced techniques such as next-generation sequencing, the nanomachine operated without the need for RNA extraction or PCR, ensuring simplicity. Having achieved ultrasensitive detection at the femtomolar level, the nanomachine outperformed the clinical biomarker CA 72-4, demonstrating superior diagnostic capabilities.
Lastly, in developing a biosensor (CPs/AuNP-AuE) for miR-100 detection, a gold nanoparticle (AuNP)-modified Au electrode (AuE) with DNA capture probes (CPs) was crafted, demonstrating enhanced electrical conductivity and an increased electrode area [59]. Differential pulse voltammetry (DPV) analysis confirmed the biosensor’s efficacy in detecting miR-100, exhibiting a linear response within a concentration range of 100 aM to 10 pM. The biosensor’s specificity was underscored by its ability to distinguish a one base-pair mistake in miR-100, and reproducibility was confirmed. When applied clinically, the biosensor revealed 100% specificity and 90% sensitivity in distinguishing miR-100 content in GC patient serum, surpassing the performance of quantitative RT-PCR.
These technologies collectively contribute to enhanced miRNA detection, painting a comprehensive picture of cellular activity and fostering improved diagnostic capabilities for EGC.
4. Exosomes-Based Nanobiosensors in Gastric Cancer
Exosomes are small extracellular vesicles, ranging in size from 30 to 150 nm with a crucial role in intercellular communication by transporting various bioactive molecules, including proteins, lipids and nucleic acids, between cells [66]. These molecules often reflect the molecular signature of the cell from which the exosome originated. As a result, exosomes can serve as carriers of specific biomarkers associated with various diseases, such as cancer. They are often released by cells early in the progression of diseases, sometimes even before clinical symptoms appear, and can be retrieved from various minimally invasive biological fluids, such as blood, urine, and saliva, making them potential indicators for early disease detection [67]. Their stability throughout the disease makes them reliable biomarkers. However, challenges like low concentrations or lengthy and complex analysis methods, which would be impractical for screening programs and resource-limited environments, have encouraged scientist to look for new solutions.
In the nanobiosensors field, exosomes can detect specific biomarkers associated with GC, such as miRs and CDs, as described in Table 3. As they carry abundant tumor-indicative information, they are considered important in liquid biopsies and have gained significant ground in early tumor nanodiagnosis due to their minimally invasive nature.
Table 3. Nanobiosensors detecting GC exosomes.
| Sensing Platform | Transducer | Biomarker | Human Sample | LoD | Takeaways | Ref. |
|---|---|---|---|---|---|---|
| MoS2 QDs-MXene heterostructure and AuNPs@biomimetic lipid layer | Electrochemical and optical (V and fluorescence) | Exosomal miR-135b | Ascites | 10 fM | Versatility, reproducibility, reliability, low background noise, high accuracy; large surface area, excellent flexibility and superior conductivity of substrates, excellent antifouling property | [68] |
| “Hot spot” AuNSs-decorated MoS2 nanocomposite (MoS2-AuNSs) aptasensors | Optical (SERS) | CD63 of exosomes | Serum | 17 particles μL−1 | Reliability, reproducibility, good stability long term, excellent Raman enhancement effect and generability in bioanalysis | [14] |
Abbreviations: MoS2—molybdenum disulfide; AuNPs—gold nanoparticles; V—voltammetry; miR—microRNA; fM—femtomolar; AuNSs—gold nanostars; SERS—surface-enhanced Raman spectroscopy; CD—cluster of differentiation.
Guo et al. created, in 2023, an innovative ECL biosensor that incorporated an MoS2 QDs-MXene heterogenous structure and excellent physicochemical properties such as a large surface-to-volume ratio and great optical features of the QDs with a AuNPs@biomimetic lipid layer [68].
Another example of a biosensing technology for the detection of exosomes is the one introduced by Pan et al., (2022), where a novel SERS nanoprobe (MoS2-AuNSs) was used to detect CD63, a representative GC exosome surface marker [14]. A 6-carboxyl-X-rhodamine (red fluorescent dye used for labelling oligonucleotides)-labelled aptamer (ROX-Apt) was used as the recognition element and was immobilized on MoS2-AuNSs, a composite material made up of molybdenum disulfide (MoS2) and gold nanospheres, providing SERS signals. The ultralow LoD aptasensor was versatile enough to detect exosomes derived from various GC cell lines. This suggests that the sensor’s performance is robust and applicable across different sources of exosomes.
The lack of standardized procedures and comprehensive validation efforts raises questions about the anticipated seamless integration of biosensors into routine clinical practices within the initially envisioned timeframe, a milestone that would have been expected by now. This nuanced perspective underscores the importance of ongoing research efforts to capitalize on the advancements achieved and bridge the existing gap between optimistic aspirations and the imperative need for comprehensive guidelines. This approach is essential to fully unlock the potential for revolutionizing early cancer diagnosis and beyond.
References
- Stomach. Globocan. 2020. Available online: https://gco.iarc.fr/today/data/factsheets/cancers/7-Stomach-fact-sheet.pdf (accessed on 15 November 2023).
- Xu, Z.-Q.; Broza, Y.Y.; Ionsecu, R.; Tisch, U.; Ding, L.; Liu, H.; Song, Q.; Pan, Y.-Y.; Xiong, F.-X.; Gu, K.-S.; et al. A nanomaterial-based breath test for distinguishing gastric cancer from benign gastric conditions. Br. J. Cancer 2013, 108, 941–950.
- Verdecchia, A.; Francisci, S.; Brenner, H.; Gatta, G.; Micheli, A.; Mangone, L.; Kunkler, I.; EUROCARE-4 Working Group. Recent cancer survival in Europe: A 2000–2002 period analysis of EUROCARE-4 data. Lancet Oncol. 2007, 8, 784–796.
- Zhang, Z.; Liu, Y.; Liu, P.; Yang, L.; Jiang, X.; Luo, D.; Yang, D. Non-invasive detection of gastric cancer relevant d -amino acids with luminescent DNA/silver nanoclusters. Nanoscale 2017, 9, 19367–19373.
- Roukos, D.H. Current status and future perspectives in gastric cancer management. Cancer Treat. Rev. 2000, 26, 243–255.
- Necula, L.; Matei, L.; Dragu, D.; I Neagu, A.; Mambet, C.; Nedeianu, S.; Bleotu, C.; Diaconu, C.C.; Chivu-Economescu, M. Recent advances in gastric cancer early diagnosis. World J. Gastroenterol. 2019, 25, 2029–2044.
- Thrift, A.P.; El-Serag, H.B. Burden of Gastric Cancer. Clin. Gastroenterol. Hepatol. 2020, 18, 534–542.
- Ferro, A.; Peleteiro, B.; Malvezzi, M.; Bosetti, C.; Bertuccio, P.; Levi, F.; Negri, E.; La Vecchia, C.; Lunet, N. Worldwide trends in gastric cancer mortality (1980–2011), with predictions to 2015, and incidence by subtype. Eur. J. Cancer 2014, 50, 1330–1344.
- Hofmarcher, T.; Lindgren, P. Cost of All Digestive Cancers in Europe Exceeds 40 Billion Euro. Digestive Cancers Europe, 14 October 2020. Available online: https://digestivecancers.eu/new-study-cost-of-all-digestive-cancers-in-europe-exceeds-40-billion-euro/ (accessed on 15 November 2023).
- The Economic Burden of Digestive Cancers in Europe. Digestive Cancers Europe. 2020. Available online: https://europacolonpolska.pl/wp-content/uploads/2020/10/DICE_WhitePaper_HealthEcoStudy_FINAL.pdf (accessed on 15 November 2023).
- Li, M.; Jiang, F.; Xue, L.; Peng, C.; Shi, Z.; Zhang, Z.; Li, J.; Pan, Y.; Wang, X.; Feng, C.; et al. Recent Progress in Biosensors for Detection of Tumor Biomarkers. Molecules 2022, 27, 7327.
- Pimenta-Melo, A.R.; Monteiro-Soares, M.; Libânio, D.; Dinis-Ribeiro, M. Missing rate for gastric cancer during upper gastrointestinal endoscopy: A systematic review and meta-analysis. Eur. J. Gastroenterol. Hepatol. 2016, 28, 1041–1049.
- Vincze, Á. Endoscopic diagnosis and treatment in gastric cancer: Current evidence and new perspectives. Front. Surg. 2023, 10, 1122454.
- Pan, H.; Dong, Y.; Gong, L.; Zhai, J.; Song, C.; Ge, Z.; Su, Y.; Zhu, D.; Chao, J.; Su, S.; et al. Sensing gastric cancer exosomes with MoS2-based SERS aptasensor. Biosens. Bioelectron. 2022, 215, 114553.
- Rød, A.M.K.; Harkestad, N.; Jellestad, F.K.; Murison, R. Comparison of commercial ELISA assays for quantification of corticosterone in serum. Sci. Rep. 2017, 7, 6748.
- Dooley, C.P. Double-Contrast Barium Meal and Upper Gastrointestinal Endoscopy: A Comparative Study. Ann. Intern. Med. 1984, 101, 538.
- Wu, J.; Fu, Z.; Yan, F.; Ju, H. Biomedical and clinical applications of immunoassays and immunosensors for tumor markers. TrAC Trends Anal. Chem. 2007, 26, 679–688.
- Wang, H.; Jin, W.; Wan, C.; Zhu, C. Diagnostic value of combined detection of CA72-4, CA19-9, and carcinoembryonic antigen comparing to CA72-4 alone in gastric cancer: A systematic review and meta-analysis. Transl. Cancer Res. 2022, 11, 848–856.
- Liou, J.-M.; Malfertheiner, P.; Lee, Y.-C.; Sheu, B.-S.; Sugano, K.; Cheng, H.-C.; Yeoh, K.-G.; Hsu, P.-I.; Goh, K.-L.; Mahachai, V.; et al. Screening and eradication of Helicobacter pylori for gastric cancer prevention: The Taipei global consensus. Gut 2020, 69, 2093–2112.
- Heo, Y.J.; Kim, B.; Kim, H.; Kim, S.; Jang, M.S.; Kim, K.-M. PD-L1 expression in paired biopsies and surgical specimens in gastric adenocarcinoma: A digital image analysis study. Pathol.—Res. Pract. 2021, 218, 153338.
- Go, S.I.; Ko, G.H.; Lee, W.S.; Kim, R.B.; Lee, J.H.; Jeong, S.H.; Lee, Y.J.; Hong, S.C.; Ha, W.S. CD44 Variant 9 Serves as a Poor Prognostic Marker in Early Gastric Cancer, But Not in Advanced Gastric Cancer. Cancer Res. Treat. 2016, 48, 142–152.
- Umapathy, V.R.; Natarajan, P.M.; Swamikannu, B.; Moses, J.; Jones, S.; Chandran, M.P.; Anbumozhi, M.K. Emerging Biosensors for Oral Cancer Detection and Diagnosis—A Review Unravelling Their Role in Past and Present Advancements in the Field of Early Diagnosis. Biosensors 2022, 12, 498.
- Wang, Z.; Fan, Y.; Chen, J.; Guo, Y.; Wu, W.; He, Y.; Xu, L.; Fu, F. A microfluidic chip-based fluorescent biosensor for the sensitive and specific detection of label-free single-base mismatch via magnetic beads-based ‘sandwich’ hybridization strategy. Electrophoresis 2013, 34, 2177–2184.
- Li, X.; Ai, S.; Lu, X.; Liu, S.; Guan, W. Nanotechnology-based strategies for gastric cancer imaging and treatment. RSC Adv. 2021, 11, 35392–35407.
- Zhang, Y.; Li, M.; Gao, X.; Chen, Y.; Liu, T. Nanotechnology in cancer diagnosis: Progress, challenges and opportunities. J. Hematol. Oncol. J. Hematol. Oncol. 2019, 12, 137.
- Haleem, A.; Javaid, M.; Singh, R.P.; Suman, R.; Rab, S. Biosensors applications in medical field: A brief review. Sens. Int. 2021, 2, 100100.
- Yuwen, L.; Zhang, S.; Chao, J. Recent Advances in DNA Nanotechnology-Enabled Biosensors for Virus Detection. Biosensors 2023, 13, 822.
- Naresh, V.; Lee, N. A Review on Biosensors and Recent Development of Nanostructured Materials-Enabled Biosensors. Sensors 2021, 21, 1109.
- Soroka, M.; Wasowicz, B.; Rymaszewska, A. Loop-Mediated Isothermal Amplification (LAMP): The Better Sibling of PCR? Cells 2021, 10, 1931.
- Ge, S.; Ran, M.; Mao, Y.; Sun, Y.; Zhou, X.; Li, L.; Cao, X. A novel DNA biosensor for the ultrasensitive detection of DNA methyltransferase activity based on a high-density ‘hot spot’ SERS substrate and rolling circle amplification strategy. Analyst 2021, 146, 5326–5336.
- Rahman, M.; Niu, J.; Cui, X.; Zhou, C.; Tang, N.; Jin, H.; Cui, D. Electrochemical Biosensor Based on l -Arginine and rGO-AuNSs Deposited on the Electrode Combined with DNA Probes for Ultrasensitive Detection of the Gastric Cancer-Related PIK3CA Gene of ctDNA. ACS Appl. Bio Mater. 2022, 5, 5094–5103.
- Yu, Z.; Gopinath, S.C.B.; Lakshmipriya, T.; Anbu, P. Single-walled carbon nanotube-gold urchin nanohybrid for identifying gastric cancer on dimicroelectrodes junction. J. Taiwan Inst. Chem. Eng. 2021, 121, 108–114.
- Huang, C.; Hu, S.; Zhang, X.; Cui, H.; Wu, L.; Yang, N.; Zhou, W.; Chu, P.K.; Yu, X.-F. Sensitive and selective ctDNA detection based on functionalized black phosphorus nanosheets. Biosens. Bioelectron. 2020, 165, 112384.
- Guthula, L.S.; Yeh, K.-T.; Huang, W.-L.; Chen, C.-H.; Chen, Y.-L.; Huang, C.-J.; Chau, L.-K.; Chan, M.W.; Lin, S.-H. Quantitative and amplification-free detection of SOCS-1 CpG methylation percentage analyses in gastric cancer by fiber optic nanoplasmonic biosensor. Biosens. Bioelectron. 2022, 214, 114540.
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38.
- Delpu, Y.; Cordelier, P.; Cho, W.C.; Torrisani, J. DNA Methylation and Cancer Diagnosis. Int. J. Mol. Sci. 2013, 14, 15029–15058.
- Dammann, R.H.; Richter, A.M.; Jiménez, A.P.; Woods, M.; Küster, M.; Witharana, C. Impact of Natural Compounds on DNA Methylation Levels of the Tumor Suppressor Gene RASSF1A in Cancer. Int. J. Mol. Sci. 2017, 18, 2160.
- Das, G.M.; Managò, S.; Mangini, M.; De Luca, A.C. Biosensing Using SERS Active Gold Nanostructures. Nanomaterials 2021, 11, 2679.
- Pérez-Jiménez, A.I.; Lyu, D.; Lu, Z.; Liu, G.; Ren, B. Surface-enhanced Raman spectroscopy: Benefits, trade-offs and future developments. Chem. Sci. 2020, 11, 4563–4577.
- Cao, X.; Ge, S.; Hua, W.; Zhou, X.; Lu, W.; Gu, Y.; Li, Z.; Qian, Y. A pump-free and high-throughput microfluidic chip for highly sensitive SERS assay of gastric cancer-related circulating tumor DNA via a cascade signal amplification strategy. J. Nanobiotechnology 2022, 20, 271.
- Cheng, F.; Su, L.; Qian, C. Circulating tumor DNA: A promising biomarker in the liquid biopsy of cancer. Oncotarget 2016, 7, 48832–48841.
- Liquid Biopsy: The Value of Different Bodily Fluids—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/35073730/ (accessed on 8 January 2024).
- Ma, W.; Du, H.; Zhang, M.; Mori, J.; Ren, X.; Wang, H.; Zhang, X. One-Step Synthesis of Tunable Zinc-Based Nanohybrids as an Ultrasensitive DNA Signal Amplification Platform. ACS Appl. Mater. Interfaces 2020, 12, 2983–2990.
- Dang, D.K.; Park, B.H. Circulating tumor DNA: Current challenges for clinical utility. J. Clin. Investig. 2022, 132, e154941.
- Bittla, P.; Kaur, S.; Sojitra, V.; Zahra, A.; Hutchinson, J.; Folawemi, O.; Khan, S. Exploring Circulating Tumor DNA (CtDNA) and Its Role in Early Detection of Cancer: A Systematic Review. Cureus 2023, 15, e45784.
- He, B.; Zhao, Z.; Cai, Q.; Zhang, Y.; Zhang, P.; Shi, S.; Xie, H.; Peng, X.; Yin, W.; Tao, Y.; et al. miRNA-based biomarkers, therapies, and resistance in Cancer. Int. J. Biol. Sci. 2020, 16, 2628–2647.
- Rezayi, M.; Farjami, Z.; Hosseini, Z.S.; Ebrahimi, N.; Abouzari-Lotf, E. MicroRNA-based Biosensors for Early Detection of Cancers. Curr. Pharm. Des. 2019, 24, 4675–4680.
- Pallares, R.M.; Thanh, N.T.K.; Su, X. Sensing of circulating cancer biomarkers with metal nanoparticles. Nanoscale 2019, 11, 22152–22171.
- Radfar, S.; Ghanbari, R.; Isfahani, A.A.; Rezaei, H.; Kheirollahi, M. A novel signal amplification tag to develop rapid and sensitive aptamer-based biosensors. Bioelectrochemistry 2022, 145, 108087.
- Daneshpour, M.; Omidfar, K.; Ghanbarian, H. A novel electrochemical nanobiosensor for the ultrasensitive and specific detection of femtomolar-level gastric cancer biomarker miRNA-106a. Beilstein J. Nanotechnol. 2016, 7, 2023–2036.
- Daneshpour, M.; Karimi, B.; Omidfar, K. Simultaneous detection of gastric cancer-involved miR-106a and let-7a through a dual-signal-marked electrochemical nanobiosensor. Biosens. Bioelectron. 2018, 109, 197–205.
- Zhai, J.; Li, X.; Zhang, J.; Pan, H.; Peng, Q.; Gan, H.; Su, S.; Yuwen, L.; Song, C. SERS/electrochemical dual-mode biosensor based on multi-functionalized molybdenum disulfide nanosheet probes and SERS-active Ag nanorods array electrodes for reliable detection of cancer-related miRNA. Sens. Actuators B Chem. 2022, 368, 132245.
- Shahbazi-Derakhshi, P.; Mahmoudi, E.; Majidi, M.M.; Sohrabi, H.; Amini, M.; Majidi, M.R.; Niaei, A.; Shaykh-Baygloo, N.; Mokhtarzadeh, A. An Ultrasensitive miRNA-Based Genosensor for Detection of MicroRNA 21 in Gastric Cancer Cells Based on Functional Signal Amplifier and Synthesized Perovskite-Graphene Oxide and AuNPs. Biosensors 2023, 13, 172.
- Wang, Z.; Xue, Z.; Hao, X.; Miao, C.; Zhang, J.; Zheng, Y.; Zheng, Z.; Lin, X.; Weng, S. Ratiometric fluorescence sensor based on carbon dots as internal reference signal and T7 exonuclease-assisted signal amplification strategy for microRNA-21 detection. Anal. Chim. Acta 2020, 1103, 212–219.
- Li, B.; Liu, F.; Peng, Y.; Zhou, Y.; Fan, W.; Yin, H.; Ai, S.; Zhang, X. Two-stage cyclic enzymatic amplification method for ultrasensitive electrochemical assay of microRNA-21 in the blood serum of gastric cancer patients. Biosens. Bioelectron. 2016, 79, 307–312.
- Li, M.; Li, Z.; Wang, P.; Ma, Q. A novel bimetallic MXene derivative QD-based ECL sensor for miRNA-27a-3p detection. Biosens. Bioelectron. 2023, 228, 115225.
- Li, Z.; Wang, P.; Liang, Z.; Wang, D.; Nie, Y.; Ma, Q. Bismuth Nano-Nest/Ti3CN Quantum Dot-Based Surface Plasmon Coupling Electrochemiluminescence Sensor for Ascites miRNA-421 Detection. Anal. Chem. 2023, 95, 9706–9713.
- Zhang, P.; Tong, Y.; Huang, X.; Chen, Y.; Li, Y.; Luan, D.; Li, J.; Wang, C.; Li, P.; Du, L.; et al. The Dual-Response–Single-Amplification Fluorescent Nanomachine for Tumor Imaging and Gastric Cancer Diagnosis. ACS Nano 2023, 17, 16553–16564.
- Zhuang, J.; Wan, H.; Zhang, X. Electrochemical detection of miRNA-100 in the sera of gastric cancer patients based on DSN-assisted amplification. Talanta 2021, 225, 121981.
- Pan, Y.-J.; Zhuang, Y.; Zheng, J.-N.; Pei, D.-S. MiR-106a: Promising biomarker for cancer. Bioorg. Med. Chem. Lett. 2016, 26, 5373–5377.
- Park, S.-H.; Lee, J.; Yeo, J.-S. On-chip plasmonic detection of microRNA-106a in gastric cancer using hybridized gold nanoparticles. Sens. Actuators B Chem. 2018, 262, 703–709.
- Shafiee, M.R.M.; Parhizkar, J. Au nanoparticles/g-C3N4 modified biosensor for electrochemical detection of gastric cancer miRNA based on hairpin locked nucleic acids probe. Nanomedicine Res. J. 2020, 5, 152–159.
- Kumar, V.; Guleria, P. Application of DNA-Nanosensor for Environmental Monitoring: Recent Advances and Perspectives. Curr. Pollut. Rep. 2020.
- Sekar, D.; Krishnan, R.; Thirugnanasambantham, K.; Rajasekaran, B.; Islam, V.I.H.; Sekar, P. Significance of microRNA 21 in gastric cancer. Clin. Res. Hepatol. Gastroenterol. 2016, 40, 538–545.
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to biosensors. Essays Biochem. 2016, 60, 1–8.
- Chen, H.; Wang, L.; Zeng, X.; Schwarz, H.; Nanda, H.S.; Peng, X.; Zhou, Y. Exosomes, a New Star for Targeted Delivery. Front. Cell Dev. Biol. 2021, 9, 751079.
- Lv, Z.; Fu, K.; Zhang, Q. Advances of exosomes-based applications in diagnostic biomarkers for dental disease and dental regeneration. Colloids Surf. B Biointerfaces 2023, 229, 113429.
- Guo, Y.; Nie, Y.; Wang, P.; Li, Z.; Ma, Q. MoS2 QDs-MXene heterostructure-based ECL sensor for the detection of miRNA-135b in gastric cancer exosomes. Talanta 2023, 259, 124559.
More
Information
Subjects:
Oncology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
575
Revisions:
2 times
(View History)
Update Date:
20 Mar 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




