Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Serigne Fallou Wade | -- | 3020 | 2024-03-16 05:32:25 | | | |
| 2 | Mona Zou | + 1 word(s) | 3021 | 2024-03-18 10:11:59 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Wade, S.F.; Diouara, A.A.M.; Ngom, B.; Thiam, F.; Dia, N. SARS-CoV-2 and Other Respiratory Viruses in Olfactory Pathophysiology. Encyclopedia. Available online: https://encyclopedia.pub/entry/56353 (accessed on 07 February 2026).
Wade SF, Diouara AAM, Ngom B, Thiam F, Dia N. SARS-CoV-2 and Other Respiratory Viruses in Olfactory Pathophysiology. Encyclopedia. Available at: https://encyclopedia.pub/entry/56353. Accessed February 07, 2026.
Wade, Serigne Fallou, Abou Abdallah Malick Diouara, Babacar Ngom, Fatou Thiam, Ndongo Dia. "SARS-CoV-2 and Other Respiratory Viruses in Olfactory Pathophysiology" Encyclopedia, https://encyclopedia.pub/entry/56353 (accessed February 07, 2026).
Wade, S.F., Diouara, A.A.M., Ngom, B., Thiam, F., & Dia, N. (2024, March 16). SARS-CoV-2 and Other Respiratory Viruses in Olfactory Pathophysiology. In Encyclopedia. https://encyclopedia.pub/entry/56353
Wade, Serigne Fallou, et al. "SARS-CoV-2 and Other Respiratory Viruses in Olfactory Pathophysiology." Encyclopedia. Web. 16 March, 2024.
Copy Citation
Acute respiratory viruses (ARVs) are the leading cause of diseases in humans worldwide. High-risk individuals, including children and the elderly, could potentially develop severe illnesses that could result in hospitalization or death in the worst case. The most common ARVs are the Human respiratory syncytial virus, Human Metapneumovirus, Human Parainfluenza Virus, rhinovirus, coronaviruses (including SARS and MERS CoV), adenoviruses, Human Bocavirus, enterovirus (-D68 and 71), and influenza viruses. The olfactory deficits due to ARV infection are a common symptom among patients.
respiratory viruses
anosmia
olfaction disorders
loss of smell
COVID-19
1. Viral Infection Causing Olfactory Dysfunction
The fact that the olfactory receptor neurons (ORNs) are found in the nasal cavity and expressed in the OE makes them directly exposed to all kinds of air-bound and air-way pathogens that make the ORNs vulnerable. Whether the cause is physiological or pathological, the lifespan of ORNs is relatively short with a few weeks in the OE. Moreover, stem cell reprogramming ensures the continuous regeneration of new ORNs from OE basal cells either in a physiological turnover of ORNs or in response to inflammation and OE severe damage mediated by neural injury [1][2][3]. Several airway pathogens, such as viruses, are causing damage to OE, particularly through the sustentacular cells, triggering anosmia, hyposmia, phantosmia, or parmosmia in mammals [4][5][6][7][8]. Many respiratory tract infections due to viruses like RV, PIV, RSV, CoV, and Epstein-Barr viruses (EBV) [5][9][10][11][12] have been involved in the development of olfactory disorders such as partial or total loss of smell. Doty and others have termed this pathology as virus-induced olfactory dysfunction (PVOD) [13][14][15][16]. Viral infection destroys many cells within the apical layer of the OE, which could lead to ORN functional impairment in the nose. Interestingly, the OE basal cells can constantly replace damaged ORNs with new olfactory neurons, allowing patients to recover functional olfactory responses [2][3]. In the following sections, the common viral infection of the URT leading to olfactory dysfunction like anosmia, hyposmia, phantosmia, and parmosmia [9] in animal models and humans will be discussed.
2. Viruses Impacting Respiratory System
The respiratory system is exposed to the environment and is in permanent contact with air-way pathogens like viruses. A recent investigation in humans has identified 18 viruses in patients with PVOD. Several known viruses are associated with olfactory impairment, and it is crucial to investigate the mechanisms of infection as well as the specific receptors each virus targets.
2.1. Case of the Parainfluenza Viruses
The presence of parainfluenza type 3 (PIV3) was observed in human nasal epithelial cells (HNECs) from 88% of patients with PVOD as compared to 9% of control patients [17]. This data suggests the potential involvement of PIV3 infection in the upper airway pathology. PIV3 in the turbinate epithelial cells of PVOD is responsible for 60% of hyposmia and 40% of anosmia in patients [17]. PIV3 has been shown to infect the HNECs and, therefore, exacerbate the production of IFN-γ and pro-inflammatory cytokines [18]. This data is in line with a previous study suggesting that PIV3 may cause olfactory dysfunction through mechanisms other than nasal obstruction in patients [5].
2.2. Case of the Sendai Virus (SeV) and Possible Interaction with PIV
The SeV, the murine counterpart of the human PIV, has been shown to directly infect the mouse brain via the olfactory neurons [19]. Another investigation demonstrated that SeV infection led to impairing mouse olfaction. Interestingly, the virus persists in OE and OB tissues for over two months and reduces the regenerative power and functionality of the ORNs [20]. Very recent findings have shown that the depletion of nasal cilia via the silencing of CEP83, a protein critical for motile cilia formation in all ciliated cells, would impede the PIV infection. Indeed, the PIV receptor CX3CR1 colocalizes to motile cilia and is a plausible viral entry mechanism into the cells [21]. An additional study showed that the intercellular adhesion molecule-1 (ICAM-1) and related cytokine molecules are induced by PIV3, and it is thought that this activation participates in the inflammation during infection by viruses [22][23] (Figure 1). Further research using the power of the transcriptomic analysis is necessary to help delineate the role of the PIV and implicated mechanisms in the development of the broad range of olfactory dysfunctions in patients and to find host-response transcript signatures for possible treatments. The study of the entire RNA transcripts in a biological sample is referred to as transcriptomics, technically available as microarrays and RNA sequencing (RNA-seq) [24]. Only recently, some precision medicine trials using transcriptome analysis have been applied in the field of cancer [25][26][27][28][29]. Data from the study of Rodon and colleagues suggest that clinical trials using transcriptomics analysis can increase the number of patients matched to drugs [28]. Recent findings on the molecular basis of neuroimmune responses revealed that the transcriptomic results are in line with a few previously reported studies of respiratory viral infections like Influenza A virus, RV, and RSV. Moreover, these data highlighted putative biomarkers of interest as a direct reflection of each virus infection and deserve further investigation for the evaluation/prediction of future innovative treatments [30][31][32].
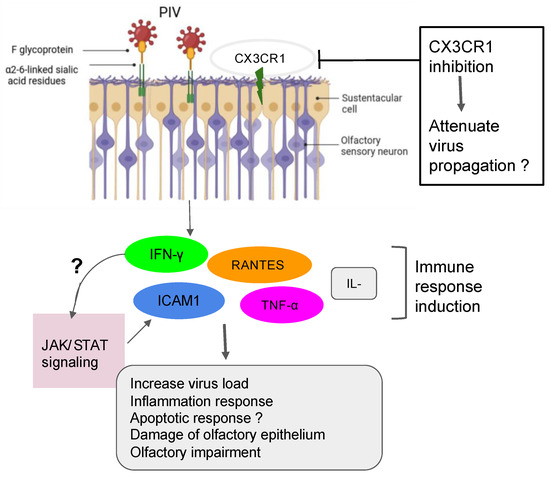
Figure 1. Action of PIV in the olfactory impairment. PIV induced the inflammation response with damage to olfactory epithelium and the impairment of olfactory response, leading to more virus susceptibility.
2.3. Case of the Respiratory Syncytial Virus (RSV)
Recent findings have demonstrated that the RSV can infect olfactory sensory neurons (OSNs) in the nasal cavity of the mouse before accessing the central nervous system of the animal [33]. As a PIV, the RSV targets the cilia of the epithelial cells in the airways by fusionning its F-glycoprotein to the cellular receptor human nucleolin (NCL). RSV also uses another mechanism that activates protein kinases like IGF1R to get into the cells [22][34][35]. Furthermore, recent studies have shown the essential role of the nucleolin RNA binding domain RBD1,2 in allowing the infection of RSV [36]. Previous works have pointed out the importance of ORN progenitors in the turnover. They showed that RSV infection causes SOX2+ ORN progenitors damage prior to the manifestation of ORN impairment. Unfortunately, the airway allergy seems to amplify this damage induced by the RSV infection, leading to a possible loss of OMP+ ORNs [37]. Transcriptomic analysis demonstrated that olfactory signaling is among the altered pathways in patients suffering from RSV infection. This finding further supports previous work that described RSV as a causative agent of post-viral olfactory dysfunction. Interestingly, the authors highlighted that this molecular signaling could be a promising future route to investigate drug targets against RSV infection [13][38]. Sourimant et al. have recently shown that 4′-fluorouridine, a ribonucleoside analog, inhibits RSV in a selective manner in cells and human airway epithelia organoids. Although this oral therapeutic drug is efficient in small animal models, several years of investigations are needed before predicting/evaluating the outcome of applying such a type of molecule in humans [39] (Figure 2). Furthermore, many efforts are underway in the phase of clinical trials when mRNA vaccines are combined with antigens to fight multiple respiratory viruses, including the RSV [40][41].
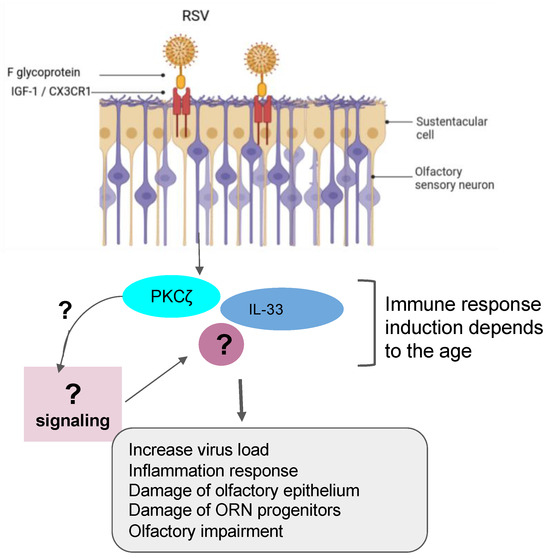
Figure 2. Role of RSV in olfactory impairment. RSV induced the inflammation response related to the age group with damage of olfactory epithelium and OSN progenitors and the impairment of olfactory response, leading to more virus susceptibility.
Recently, RVs were shown to be among the more predominant causative agents of PVOD in patients. The study showed that patients with anosmia were higher than those with hyposmia (58.8% vs. 19.0%, p = 0.018) [10]. In line with these findings, it is tempting to suggest that the persistence of the virus could be one factor that governs a more severe injury to the olfactory system. In fact, RVs primarily invade the ciliated respiratory epithelial cells via the glycoprotein members such as the intercellular adhesion molecule 1 (ICAM-1), the low-density lipoprotein receptor (LDLR) family members, and the cadherin-related family member 3 (CDHR3) [42][43][44][45]. RV infection induces Toll-like receptor 7 (TLR7) and retinoic acid-inducible gene I (RIG-1) that trigger the activation of cytokine expression (type I and type III IFNs) [46][47][48]. Understanding the mechanisms of RV viral-induced asthma for new therapeutic directions has gained more attention in the recent past [49][50][51]. Papi et al. have investigated the role of reducing agents such as DMSO on RV infection in the nasal epithelium. They showed that rhinovirus-induced ICAM-1 mRNA expression was inhibited by reducing agents in a dose-dependent manner. Interestingly, NF-κB and TNF-α activation, which is necessary for the ICAM-1 promoter, was completely abolished in those treated epithelial cells [52]. Moreover, it has been shown that CDHR3 genetic variants impact on the severity of RV-related pediatric respiratory tract infections by upregulating the epithelial expression of RV receptors, thus helping clinicians predict the susceptibility and severity of RV infection [53]. Complementary recent studies have demonstrated that both vitamin D and hydrogen peroxide play a critical role in attenuating the RV, mediating the ICAM-1 activation and the production of type I (IFN-β) and type III (IFN- λ1 and λ2) interferons respectively [54][55] (Figure 3). To further support these collective findings, it would be interesting to document transcriptomic profiles from animal and cellular models infected by RVs and treated by those molecules. These works would shed light on the importance of genes, including the oxidant biomarkers to be considered for the future in the development of the treatment against RV infection.
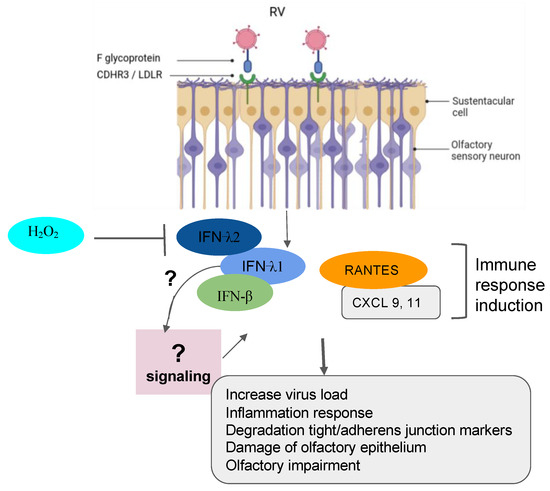
Figure 3. Implication of RV in olfactory impairment. RV induced the inflammation response with damage to the olfactory epithelium, the degradation of the tight junction and adherent junction markers, and the impairment of olfactory response leading to more virus susceptibility.
3. Mechanisms of SARS-CoV-2 Mediating the Loss of Smell
The post-COVID and the long-term-COVID have both tremendously triggered a lot of complications in different human systems. The loss or reduction of smell, among other complications of the nervous system, is an associated symptom for patients affected by different variants of COVID-19, including the omicron variant [56][57][58][59][60]. Moreover, studies reported that the prevalence of olfactory dysfunction differs greatly between populations and approaches [60][61][62]. Currently, many COVID-19 vaccines are authorized to help protect and eliminate the virus. The COVID-19 pathology and the cellular mechanism by which the olfactory dysfunction occurs have gained a lot of attention since the pandemic, and researchers are still investigating underlying signaling and complications [21][60][63][64][65][66][67][68]. Earlier in the pandemic, reports hypothesized that five potential mechanisms were considered to get insights into the olfactory dysfunction in COVID-19 patients: (1) obstruction/congestion and rhinorrhea of the nasal airway, (2) damage and loss of ORNs, (3) Olfactory center damage in the brain, (4) damage of the olfactory supporting cells in the OE, and (5) Inflammation-related olfactory epithelium dysfunction [61][69]. Butowt et al. have recently reviewed that at least the following hypotheses (1)–(3) turned out to be implausible in explaining the olfactory dysfunction in patients [69]. This allegation is further confirmed by very recent studies showing that SARS-CoV-2 infection significantly increased the expression of interferon-stimulated and inflammatory genes. Alteration of extracellular matrix genes was also observed in ALI and iALI-infected cells [64]. Here, the researchers will particularly review the mechanisms related to the second, the fourth, and the fifth scenarios according to the available findings. Healthy sensory cilia of ORNs in the olfactory epithelium are crucial in perceiving odorant molecules before sending the information to the olfactory bulbs and then to the upper parts of the brain [1]. It has been reported in humans that SARS-CoV-2 may indirectly affect the olfactory cilia, hindering the smelling system’s efficacy [70]. Reports suggested that ORNs lack the ability to express the entry proteins of SARS-CoV-2 in the OE. The virus seems to establish first contact in human nasal epithelia by binding its spike S protein to specific cells in the OE [71]. These reports are confirmed by a study based on in silico data, predicting that mature ORNs do not express the virus entry protein, the angiotensin-converting enzyme 2 (ACE2), and therefore are not likely to be infected by SARS-CoV-2 [72]. Furthermore, supporting data by Bryche et al. showed that SARS-CoV-2 was not detected in the ORNs of golden Siryan hamsters [73]. However, in a few cases, authors suggested that SARS-CoV-2 could infect ORNs in hamsters [67]. Based on the fact that COVID-19-related loss of smell disappeared within 1–2 weeks, while the regeneration of dead ORNs needs more than two weeks, many data tend to conclude that COVID-19-related olfactory dysfunction (OD) is not directly associated with the impairment of the ORNs [61][69][71][74][75]. Consequently, studying the entry protein expression within the cells in the OE will help to understand the sensitivity of the OE to SARS-CoV-2 infection related to the high prevalence of ODs in patients. Many groups are now interested in the organization of the sustentacular cells in the OE and thought that they might play a central role in leading to OD. A high level of expression of ACE2 and the transmembrane serine protease 2 (TMPRSS2) is particularly found on the sustentacular cells, suggesting a path to the neurotropism of SARS-CoV-2 in the OE. The ACE2 and TMPRSS2 are respectively known as the SARS-CoV-2 receptor and the SARS-CoV-2 cell entry-priming protease. ACE2 is found mainly on different parts of the sustentacular cells, both in humans and mice. The ACE2 and TMPRSS2 genes tend to be co-regulated [69][76][77][78][79][80]. Different approaches using tissues, cells, and organ systems in humans, golden Syrian hamsters, and hACE2 transgenic mice have been employed to study the pathological impact of the SARS-CoV-2. Here, the researchers discussed findings related particularly to the OE in inducing ODs in humans. The spike protein (S protein) of SARS-CoV-2 mediates the passage of the virus into the host cell by fusing the viral and host cell membranes. In fact, via his spike S, SARS-CoV-2 employs ACE2 as the host functional receptor and TMPRSS2 as the cellular priming protease facilitating viral uptake, both signalings being confirmed by Single-cell RNA sequencing (scRNA-seq) datasets from the Human Cell Atlas consortium [81][82][83]. Another study showed that SARS-CoV-2 Nucleocapsid protein (NP) was observed in human OE through the neuronal marker Tuj1 9 h post-infection. This data further supported the enrichment of ACE2 in human olfactory sustentacular cells [77][84]. Earlier in the pandemic, the golden Syrian hamster was used as a model to document the pathology of SARS-CoV-2 in the OE post-infection. Reports showed that the sustentacular cells are rapidly infected by SARS-CoV-2. This viral neurotropism is associated with a massive recruitment of immune cells in the OE and lamina propria, which could drive the disorganization of the OE structure [73]. This study is consistent with a high level of Tumor Necrosis Factor α (TNF α) observed in OE samples from COVID-19-suffering patients and in ALI and iALI-infected cells [64][85]. Furthermore, the inflammation induced by SARS-CoV-2 infected supporting cells may play an important role in the onset and persistence of loss of smell in patients. This SARS-CoV-2-associated inflammation status was confirmed by the transcriptome of the in vitro human airway epithelium and by analyzing the expression of selected targets in the olfactory bulb using RNA-seq and RT-qPCR tools. Interestingly, this study showed that the proinflammatory markers, including NFKBIA, CSF1, FOSL1, Cxcl10, Il-1β, Ccl5, and Irf7 overexpression continued up to 14 dpi when animals had recovered from ageusia/anosmia [64][86]. These findings are in line with a very recent study showing the implication of immune cell infiltration and altered gene expression in OE in driving persistent smell loss in a subset of patients with SARS-CoV-2. Moreover, this study particularly demonstrates that T cell-mediated inflammation lasts longer in the OE after the acute SARS-CoV-2 infection has been eliminated from the tissue, suggesting a mechanistic insight into the long-term post-COVID-19 smell loss [59]. The OE disorganization is followed by a drastic deterioration of the cilia layer of the ORNs that leads to the impairment of the olfactory capacity of the animal [73]. Investigations in humans and hamsters using, respectively, Transmission Electron Microscopy (TEM) studies and Scanning Electron Microscopy (SEM) analysis showed various levels of cilia height that undergo regeneration in the course of patient recovery, including smell restoration. Data using the golden Syrian hamster showed that the regenerated cilia in the epithelium are accompanied by a decreased expression of FOXJ1+, highlighting the importance of this marker in respiratory ciliogenesis. This later finding by Schreiner et al., could in part shed light on the inquiry of how could we regenerate cilia during patient recovery, although a lot needs to be done in the roadmap of treating loss of smell related to nasal cilia deterioration by SARS-CoV-2 [21][58][87][88] (Figure 4).
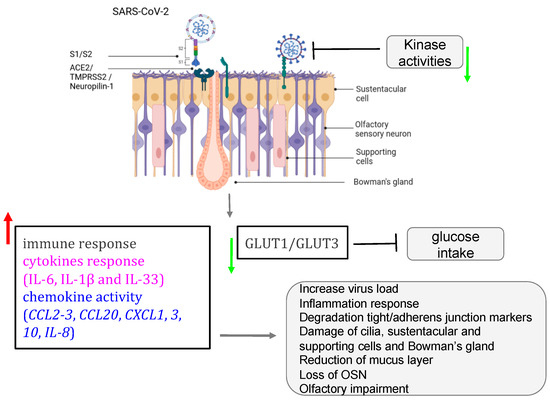
Figure 4. Role of SARS-CoV-2 in olfactory impairment. SARS-CoV-2 induced disruption of the nasal epithelium, with loss/damage of olfactory sensory neurons, sustentacular cells, Bowman’s gland, and supporting cells. All figures were created with BioRender.com (accessed on 7 October 2023).
According to the literature, different variants of SARS-CoV-2 do not directly target the ORNs in the OE; instead, they are found in the majority expressed in the sustentacular cells [6]. A recent study by Seehusen et al. showed that K18-hACE2 transgenic mouse expressing the human ACE2 is highly sensitive to at least five variants of SARS-CoV-2 that infected not only the supportive cells in OE and the respiratory epithelium but invaded the CNS of the animal five days post-infection. Interestingly, the expression of hACE2 seems to convey higher binding affinity when compared to the wild-type mouse [89]. Using this transgenic mouse is revealed to be a serious option for therapy development against loss of smell as these animals exhibited low mortality when treated with COVID-19 convalescent antisera [89][90]. For instance, the miRNA is shown to play a crucial role in the regulation of immune genes deregulated, and the development of miRNA antagonists or mimics seems to be a promising new therapeutic strategy for the treatment of patients with COVID-19 on other respiratory viruses-induced PVOD [40][41][64][91][92][93].
It is now accepted that ACE2 is not the only obligate entry for SARS-CoV-2 as it has been suggested that molecules including PIKfyve or neuropilin-1 (NRP-1) may participate in SARS-CoV-2 entry [94][95][96]. Like ACE2, NRP-1 is highly expressed in the respiratory and olfactory epithelium, which further supports the infectivity and entry of SARS-CoV-2 in the human OE. NRP-1 is not only found in supportive cells but is expressed in nearly every cell type in the nasal passages, including the ORN, therefore giving SARS-CoV-2 a route to access those cells and impair the olfactory response. Interestingly, Daly et al. demonstrated that the selective inhibition of the S1-NRP-1 interaction reduces SARS-CoV-2 infection [96][97][98].
Taken together, the high expression of ACE2, TMPRSS2, and NRP-1 in supportive and other olfactory cells and their impact on olfactory neurophysiology maintenance and in the development of human olfactory pathophysiology supports them as potential targets for signaling-based therapeutics of olfactory dysfunction.
References
- Glezer, I.; Malnic, B. Olfactory receptor function. Handb. Clin. Neurol. 2019, 164, 67–78.
- Schwob, J.E. Neural regeneration and the peripheral olfactory system. Anat. Rec. 2002, 269, 33–49.
- Graziadei, P.P.; Graziadei, G.A. Neurogenesis and neuron regeneration in the olfactory system of mammals. I. Morphological aspects of differentiation and structural organization of the olfactory sensory neurons. J. Neurocytol. 1979, 8, 1–18.
- Dicpinigaitis, P.V. Post-viral Anosmia (Loss of Sensation of Smell) Did Not Begin with COVID-19! Lung 2021, 199, 237–238.
- Suzuki, M.; Saito, K.; Min, W.P.; Vladau, C.; Toida, K.; Itoh, H.; Murakami, S. Identification of viruses in patients with postviral olfactory dysfunction. Laryngoscope 2007, 117, 272–277.
- Liang, F.; Wang, Y. COVID-19 Anosmia: High Prevalence, Plural Neuropathogenic Mechanisms, and Scarce Neurotropism of SARS-CoV-2? Viruses 2021, 13, 2225.
- Urata, S.; Maruyama, J.; Kishimoto-Urata, M.; Sattler, R.A.; Cook, R.; Lin, N.; Yamasoba, T.; Makishima, T.; Paessler, S. Regeneration Profiles of Olfactory Epithelium after SARS-CoV-2 Infection in Golden Syrian Hamsters. ACS Chem. Neurosci. 2021, 12, 589–595.
- Lee, D.Y.; Lee, W.H.; Wee, J.H.; Kim, J.W. Prognosis of postviral olfactory loss: Follow-up study for longer than one year. Am. J. Rhinol. Allergy 2014, 28, 419–422.
- Zhen Yu, L.; Luigi Angelo, V.; Paolo, B.-R.; Abigail, W.; Claire, H. Post-viral olfactory loss and parosmia. BMJ Med. 2023, 2, e000382.
- Tian, J.; Pinto, J.M.; Li, L.; Zhang, S.; Sun, Z.; Wei, Y. Identification of Viruses in Patients with Postviral Olfactory Dysfunction by Multiplex Reverse-Transcription Polymerase Chain Reaction. Laryngoscope 2021, 131, 158–164.
- Imam, S.A.; Lao, W.P.; Reddy, P.; Nguyen, S.A.; Schlosser, R.J. Is SARS-CoV-2 (COVID-19) postviral olfactory dysfunction (PVOD) different from other PVOD? World J. Otorhinolaryngol. Head. Neck Surg. 2020, 6 (Suppl. S1), S26–S32.
- Sugiura, M.; Aiba, T.; Mori, J.; Nakai, Y. An epidemiological study of postviral olfactory disorder. Acta Otolaryngol. Suppl. 1998, 538, 191–196.
- Seiden, A.M. Postviral olfactory loss. Otolaryngol. Clin. N. Am. 2004, 37, 1159–1166.
- Moran, D.T.; Jafek, B.W.; Eller, P.M.; Rowley, J.C., 3rd. Ultrastructural histopathology of human olfactory dysfunction. Microsc. Res. Tech. 1992, 23, 103–110.
- Welge-Lussen, A.; Wolfensberger, M. Olfactory disorders following upper respiratory tract infections. Adv. Otorhinolaryngol. 2006, 63, 125–132.
- Doty, R.L.; Hawkes, C.H. Chemosensory dysfunction in neurodegenerative diseases. Handb. Clin. Neurol. 2019, 164, 325–360.
- Wang, J.H.; Kwon, H.J.; Jang, Y.J. Detection of parainfluenza virus 3 in turbinate epithelial cells of postviral olfactory dysfunction patients. Laryngoscope 2007, 117, 1445–1449.
- Lewandowska-Polak, A.; Brauncajs, M.; Paradowska, E.; Jarzebska, M.; Kurowski, M.; Moskwa, S.; Lesnikowski, Z.J.; Kowalski, M.L. Human parainfluenza virus type 3 (HPIV3) induces production of IFNgamma and RANTES in human nasal epithelial cells (HNECs). J. Inflamm. 2015, 12, 16.
- Mori, I.; Komatsu, T.; Takeuchi, K.; Nakakuki, K.; Sudo, M.; Kimura, Y. Parainfluenza virus type 1 infects olfactory neurons and establishes long-term persistence in the nerve tissue. J. Gen. Virol. 1995, 76 Pt 5, 1251–1254.
- Tian, J.; Pinto, J.M.; Cui, X.; Zhang, H.; Li, L.; Liu, Y.; Wu, C.; Wei, Y. Sendai Virus Induces Persistent Olfactory Dysfunction in a Murine Model of PVOD via Effects on Apoptosis, Cell Proliferation, and Response to Odorants. PLoS ONE 2016, 11, e0159033.
- Wu, C.T.; Lidsky, P.V.; Xiao, Y.; Cheng, R.; Lee, I.T.; Nakayama, T.; Jiang, S.; He, W.; Demeter, J.; Knight, M.G.; et al. SARS-CoV-2 replication in airway epithelia requires motile cilia and microvillar reprogramming. Cell 2023, 186, 112–130 e120.
- Lee, H.S.; Volpe, S.J.; Chang, E.H. The Role of Viruses in the Inception of Chronic Rhinosinusitis. Clin. Exp. Otorhinolaryngol. 2022, 15, 310–318.
- Gao, J.; Choudhary, S.; Banerjee, A.K.; De, B.P. Human parainfluenza virus type 3 upregulates ICAM-1 (CD54) expression in a cytokine-independent manner. Gene Expr. 2000, 9, 115–121.
- Cieslik, M.; Chinnaiyan, A.M. Cancer transcriptome profiling at the juncture of clinical translation. Nat. Rev. Genet. 2018, 19, 93–109.
- Worst, B.C.; van Tilburg, C.M.; Balasubramanian, G.P.; Fiesel, P.; Witt, R.; Freitag, A.; Boudalil, M.; Previti, C.; Wolf, S.; Schmidt, S.; et al. Next-generation personalised medicine for high-risk paediatric cancer patients—The INFORM pilot study. Eur. J. Cancer 2016, 65, 91–101.
- Weidenbusch, B.; Richter, G.H.S.; Kesper, M.S.; Guggemoos, M.; Gall, K.; Prexler, C.; Kazantsev, I.; Sipol, A.; Lindner, L.; Nathrath, M.; et al. Transcriptome based individualized therapy of refractory pediatric sarcomas: Feasibility, tolerability and efficacy. Oncotarget 2018, 9, 20747–20760.
- Tsimberidou, A.M.; Fountzilas, E.; Bleris, L.; Kurzrock, R. Transcriptomics and solid tumors: The next frontier in precision cancer medicine. Semin. Cancer Biol. 2022, 84, 50–59.
- Rodon, J.; Soria, J.C.; Berger, R.; Miller, W.H.; Rubin, E.; Kugel, A.; Tsimberidou, A.; Saintigny, P.; Ackerstein, A.; Brana, I.; et al. Genomic and transcriptomic profiling expands precision cancer medicine: The WINTHER trial. Nat. Med. 2019, 25, 751–758.
- Oberg, J.A.; Glade Bender, J.L.; Sulis, M.L.; Pendrick, D.; Sireci, A.N.; Hsiao, S.J.; Turk, A.T.; Dela Cruz, F.S.; Hibshoosh, H.; Remotti, H.; et al. Implementation of next generation sequencing into pediatric hematology-oncology practice: Moving beyond actionable alterations. Genome Med. 2016, 8, 133.
- Nicolas De Lamballerie, C.; Pizzorno, A.; Dubois, J.; Padey, B.; Julien, T.; Traversier, A.; Carbonneau, J.; Orcel, E.; Lina, B.; Hamelin, M.E.; et al. Human Respiratory Syncytial Virus-Induced Immune Signature of Infection Revealed by Transcriptome Analysis of Clinical Pediatric Nasopharyngeal Swab Samples. J. Infect. Dis. 2021, 223, 1052–1061.
- Dissanayake, T.K.; Schauble, S.; Mirhakkak, M.H.; Wu, W.L.; Ng, A.C.; Yip, C.C.Y.; Lopez, A.G.; Wolf, T.; Yeung, M.L.; Chan, K.H.; et al. Comparative Transcriptomic Analysis of Rhinovirus and Influenza Virus Infection. Front. Microbiol. 2020, 11, 1580.
- Al-Shalan, H.A.M.; Hu, D.; Wang, P.; Uddin, J.; Chopra, A.; Greene, W.K.; Ma, B. Transcriptomic Profiling of Influenza A Virus-Infected Mouse Lung at Recovery Stage Using RNA Sequencing. Viruses 2023, 15, 2198.
- Bryche, B.; Fretaud, M.; Saint-Albin Deliot, A.; Galloux, M.; Sedano, L.; Langevin, C.; Descamps, D.; Rameix-Welti, M.A.; Eleouet, J.F.; Le Goffic, R.; et al. Respiratory syncytial virus tropism for olfactory sensory neurons in mice. J. Neurochem. 2020, 155, 137–153.
- Griffiths, C.D.; Bilawchuk, L.M.; McDonough, J.E.; Jamieson, K.C.; Elawar, F.; Cen, Y.; Duan, W.; Lin, C.; Song, H.; Casanova, J.L.; et al. IGF1R is an entry receptor for respiratory syncytial virus. Nature 2020, 583, 615–619.
- Tayyari, F.; Marchant, D.; Moraes, T.J.; Duan, W.; Mastrangelo, P.; Hegele, R.G. Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nat. Med. 2011, 17, 1132–1135.
- Mastrangelo, P.; Chin, A.A.; Tan, S.; Jeon, A.H.; Ackerley, C.A.; Siu, K.K.; Lee, J.E.; Hegele, R.G. Identification of RSV Fusion Protein Interaction Domains on the Virus Receptor, Nucleolin. Viruses 2021, 13, 261.
- Ueha, R.; Mukherjee, S.; Ueha, S.; de Almeida Nagata, D.E.; Sakamoto, T.; Kondo, K.; Yamasoba, T.; Lukacs, N.W.; Kunkel, S.L. Viral disruption of olfactory progenitors is exacerbated in allergic mice. Int. Immunopharmacol. 2014, 22, 242–247.
- Barral-Arca, R.; Gomez-Carballa, A.; Cebey-Lopez, M.; Bello, X.; Martinon-Torres, F.; Salas, A. A Meta-Analysis of Multiple Whole Blood Gene Expression Data Unveils a Diagnostic Host-Response Transcript Signature for Respiratory Syncytial Virus. Int. J. Mol. Sci. 2020, 21, 1831.
- Sourimant, J.; Lieber, C.M.; Aggarwal, M.; Cox, R.M.; Wolf, J.D.; Yoon, J.J.; Toots, M.; Ye, C.; Sticher, Z.; Kolykhalov, A.A.; et al. 4′-Fluorouridine is an oral antiviral that blocks respiratory syncytial virus and SARS-CoV-2 replication. Science 2022, 375, 161–167.
- Li, H.H.; Xu, J.; He, L.; Denny, L.I.; Rustandi, R.R.; Dornadula, G.; Fiorito, B.; Zhang, Z.Q. Development and qualification of cell-based relative potency assay for a human respiratory syncytial virus (RSV) mRNA vaccine. J. Pharm. Biomed. Anal. 2023, 234, 115523.
- Whitaker, J.A.; Sahly, H.M.E.; Healy, C.M. mRNA vaccines against respiratory viruses. Curr. Opin. Infect. Dis. 2023, 36, 385–393.
- Bochkov, Y.A.; Watters, K.; Ashraf, S.; Griggs, T.F.; Devries, M.K.; Jackson, D.J.; Palmenberg, A.C.; Gern, J.E. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc. Natl. Acad. Sci. USA 2015, 112, 5485–5490.
- Staunton, D.E.; Merluzzi, V.J.; Rothlein, R.; Barton, R.; Marlin, S.D.; Springer, T.A. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell 1989, 56, 849–853.
- Hofer, F.; Gruenberger, M.; Kowalski, H.; Machat, H.; Huettinger, M.; Kuechler, E.; Blaas, D. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc. Natl. Acad. Sci. USA 1994, 91, 1839–1842.
- Bochkov, Y.A.; Gern, J.E. Rhinoviruses and Their Receptors: Implications for Allergic Disease. Curr. Allergy Asthma Rep. 2016, 16, 30.
- Lavoie, T.B.; Kalie, E.; Crisafulli-Cabatu, S.; Abramovich, R.; DiGioia, G.; Moolchan, K.; Pestka, S.; Schreiber, G. Binding and activity of all human alpha interferon subtypes. Cytokine 2011, 56, 282–289.
- Jaks, E.; Gavutis, M.; Uze, G.; Martal, J.; Piehler, J. Differential receptor subunit affinities of type I interferons govern differential signal activation. J. Mol. Biol. 2007, 366, 525–539.
- Donnelly, R.P.; Sheikh, F.; Kotenko, S.V.; Dickensheets, H. The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J. Leukoc. Biol. 2004, 76, 314–321.
- Wang, Y.; Ninaber, D.K.; van Schadewijk, A.; Hiemstra, P.S. Tiotropium and Fluticasone Inhibit Rhinovirus-Induced Mucin Production via Multiple Mechanisms in Differentiated Airway Epithelial Cells. Front. Cell Infect. Microbiol. 2020, 10, 278.
- Lo, D.; Kennedy, J.L.; Kurten, R.C.; Panettieri, R.A., Jr.; Koziol-White, C.J. Modulation of airway hyperresponsiveness by rhinovirus exposure. Respir. Res. 2018, 19, 208.
- Loxham, M.; Smart, D.E.; Bedke, N.J.; Smithers, N.P.; Filippi, I.; Blume, C.; Swindle, E.J.; Tariq, K.; Howarth, P.H.; Holgate, S.T.; et al. Allergenic proteases cleave the chemokine CX3CL1 directly from the surface of airway epithelium and augment the effect of rhinovirus. Mucosal Immunol. 2018, 11, 404–414.
- Papi, A.; Papadopoulos, N.G.; Stanciu, L.A.; Bellettato, C.M.; Pinamonti, S.; Degitz, K.; Holgate, S.T.; Johnston, S.L. Reducing agents inhibit rhinovirus-induced up-regulation of the rhinovirus receptor intercellular adhesion molecule-1 (ICAM-1) in respiratory epithelial cells. FASEB J. 2002, 16, 1934–1936.
- Song, Y.P.; Tang, M.F.; Leung, A.S.Y.; Tao, K.P.; Chan, O.M.; Wong, G.W.K.; Chan, P.K.S.; Chan, R.W.Y.; Leung, T.F. Interactive effects between CDHR3 genotype and rhinovirus species for diagnosis and severity of respiratory tract infections in hospitalized children. Microbiol. Spectr. 2023, 11, e0118123.
- Lee, S.H.; Han, M.S.; Lee, T.H.; Lee, D.B.; Park, J.H.; Lee, S.H.; Kim, T.H. Hydrogen peroxide attenuates rhinovirus-induced anti-viral interferon secretion in sinonasal epithelial cells. Front. Immunol. 2023, 14, 1086381.
- Greiller, C.L.; Suri, R.; Jolliffe, D.A.; Kebadze, T.; Hirsman, A.G.; Griffiths, C.J.; Johnston, S.L.; Martineau, A.R. Vitamin D attenuates rhinovirus-induced expression of intercellular adhesion molecule-1 (ICAM-1) and platelet-activating factor receptor (PAFR) in respiratory epithelial cells. J. Steroid Biochem. Mol. Biol. 2019, 187, 152–159.
- Khani, E.; Khiali, S.; Beheshtirouy, S.; Entezari-Maleki, T. Potential pharmacologic treatments for COVID-19 smell and taste loss: A comprehensive review. Eur. J. Pharmacol. 2021, 912, 174582.
- Silva Andrade, B.; Siqueira, S.; de Assis Soares, W.R.; de Souza Rangel, F.; Santos, N.O.; Dos Santos Freitas, A.; Ribeiro da Silveira, P.; Tiwari, S.; Alzahrani, K.J.; Goes-Neto, A.; et al. Long-COVID and Post-COVID Health Complications: An Up-to-Date Review on Clinical Conditions and Their Possible Molecular Mechanisms. Viruses 2021, 13, 700.
- Fernandez-de-Las-Penas, C.; Cancela-Cilleruelo, I.; Rodriguez-Jimenez, J.; Gomez-Mayordomo, V.; Pellicer-Valero, O.J.; Martin-Guerrero, J.D.; Hernandez-Barrera, V.; Arendt-Nielsen, L.; Torres-Macho, J. Associated-Onset Symptoms and Post-COVID-19 Symptoms in Hospitalized COVID-19 Survivors Infected with Wuhan, Alpha or Delta SARS-CoV-2 Variant. Pathogens 2022, 11, 725.
- Rodriguez-Sevilla, J.J.; Guerri-Fernadez, R.; Bertran Recasens, B. Is There Less Alteration of Smell Sensation in Patients with Omicron SARS-CoV-2 Variant Infection? Front Med. 2022, 9, 852998.
- Chee, J.; Chern, B.; Loh, W.S.; Mullol, J.; Wang, Y. Pathophysiology of SARS-CoV-2 Infection of Nasal Respiratory and Olfactory Epithelia and Its Clinical Impact. Curr. Allergy Asthma Rep. 2023, 23, 121–131.
- Mutiawati, E.; Fahriani, M.; Mamada, S.S.; Fajar, J.K.; Frediansyah, A.; Maliga, H.A.; Ilmawan, M.; Emran, T.B.; Ophinni, Y.; Ichsan, I.; et al. Anosmia and dysgeusia in SARS-CoV-2 infection: Incidence and effects on COVID-19 severity and mortality, and the possible pathobiology mechanisms—A systematic review and meta-analysis. F1000Res 2021, 10, 40.
- Butowt, R.; von Bartheld, C.S. Anosmia in COVID-19: Underlying Mechanisms and Assessment of an Olfactory Route to Brain Infection. Neuroscientist 2021, 27, 582–603.
- Khan, M.; Yoo, S.J.; Clijsters, M.; Backaert, W.; Vanstapel, A.; Speleman, K.; Lietaer, C.; Choi, S.; Hether, T.D.; Marcelis, L.; et al. Visualizing in deceased COVID-19 patients how SARS-CoV-2 attacks the respiratory and olfactory mucosae but spares the olfactory bulb. Cell 2021, 184, 5932–5949 e5915.
- Assou, S.; Ahmed, E.; Morichon, L.; Nasri, A.; Foisset, F.; Bourdais, C.; Gros, N.; Tieo, S.; Petit, A.; Vachier, I.; et al. The Transcriptome Landscape of the In Vitro Human Airway Epithelium Response to SARS-CoV-2. Int. J. Mol. Sci. 2023, 24, 12017.
- Kalra, R.S.; Dhanjal, J.K.; Meena, A.S.; Kalel, V.C.; Dahiya, S.; Singh, B.; Dewanjee, S.; Kandimalla, R. COVID-19, Neuropathology, and Aging: SARS-CoV-2 Neurological Infection, Mechanism, and Associated Complications. Front. Aging Neurosci. 2021, 13, 662786.
- Rebholz, H.; Braun, R.J.; Ladage, D.; Knoll, W.; Kleber, C.; Hassel, A.W. Loss of Olfactory Function-Early Indicator for COVID-19, Other Viral Infections and Neurodegenerative Disorders. Front. Neurol. 2020, 11, 569333.
- Reyna, R.A.; Kishimoto-Urata, M.; Urata, S.; Makishima, T.; Paessler, S.; Maruyama, J. Recovery of anosmia in hamsters infected with SARS-CoV-2 is correlated with repair of the olfactory epithelium. Sci. Rep. 2022, 12, 628.
- Tanzadehpanah, H.; Lotfian, E.; Avan, A.; Saki, S.; Nobari, S.; Mahmoodian, R.; Sheykhhasan, M.; Froutagh, M.H.S.; Ghotbani, F.; Jamshidi, R.; et al. Role of SARS-CoV-2 and ACE2 in the pathophysiology of peripheral vascular diseases. Biomed. Pharmacother. 2023, 166, 115321.
- Butowt, R.; Bilinska, K.; von Bartheld, C.S. Olfactory dysfunction in COVID-19: New insights into the underlying mechanisms. Trends Neurosci. 2023, 46, 75–90.
- Buqaileh, R.; Saternos, H.; Ley, S.; Aranda, A.; Forero, K.; AbouAlaiwi, W.A. Can cilia provide an entry gateway for SARS-CoV-2 to human ciliated cells? Physiol. Genom. 2021, 53, 249–258.
- Butowt, R.; Meunier, N.; Bryche, B.; von Bartheld, C.S. The olfactory nerve is not a likely route to brain infection in COVID-19: A critical review of data from humans and animal models. Acta Neuropathol. 2021, 141, 809–822.
- Bilinska, K.; Jakubowska, P.; Von Bartheld, C.S.; Butowt, R. Expression of the SARS-CoV-2 Entry Proteins, ACE2 and TMPRSS2, in Cells of the Olfactory Epithelium: Identification of Cell Types and Trends with Age. ACS Chem. Neurosci. 2020, 11, 1555–1562.
- Bryche, B.; St Albin, A.; Murri, S.; Lacote, S.; Pulido, C.; Ar Gouilh, M.; Lesellier, S.; Servat, A.; Wasniewski, M.; Picard-Meyer, E.; et al. Massive transient damage of the olfactory epithelium associated with infection of sustentacular cells by SARS-CoV-2 in golden Syrian hamsters. Brain Behav. Immun. 2020, 89, 579–586.
- Lechien, J.R.; Chiesa-Estomba, C.M.; De Siati, D.R.; Horoi, M.; Le Bon, S.D.; Rodriguez, A.; Dequanter, D.; Blecic, S.; El Afia, F.; Distinguin, L.; et al. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): A multicenter European study. Eur. Arch. Otorhinolaryngol. 2020, 277, 2251–2261.
- Lee, Y.; Min, P.; Lee, S.; Kim, S.W. Prevalence and Duration of Acute Loss of Smell or Taste in COVID-19 Patients. J. Korean Med. Sci. 2020, 35, e174.
- Fodoulian, L.; Tuberosa, J.; Rossier, D.; Boillat, M.; Kan, C.; Pauli, V.; Egervari, K.; Lobrinus, J.A.; Landis, B.N.; Carleton, A.; et al. SARS-CoV-2 Receptors and Entry Genes Are Expressed in the Human Olfactory Neuroepithelium and Brain. iScience 2020, 23, 101839.
- Chen, M.; Shen, W.; Rowan, N.R.; Kulaga, H.; Hillel, A.; Ramanathan, M., Jr.; Lane, A.P. Elevated ACE-2 expression in the olfactory neuroepithelium: Implications for anosmia and upper respiratory SARS-CoV-2 entry and replication. Eur. Respir. J. 2020, 56, 2001948.
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280 e278.
- Ziegler, C.G.K.; Allon, S.J.; Nyquist, S.K.; Mbano, I.M.; Miao, V.N.; Tzouanas, C.N.; Cao, Y.; Yousif, A.S.; Bals, J.; Hauser, B.M.; et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 2020, 181, 1016–1035.e1019.
- Gkogkou, E.; Barnasas, G.; Vougas, K.; Trougakos, I.P. Expression profiling meta-analysis of ACE2 and TMPRSS2, the putative anti-inflammatory receptor and priming protease of SARS-CoV-2 in human cells, and identification of putative modulators. Redox Biol. 2020, 36, 101615.
- Sungnak, W.; Huang, N.; Becavin, C.; Berg, M.; Queen, R.; Litvinukova, M.; Talavera-Lopez, C.; Maatz, H.; Reichart, D.; Sampaziotis, F.; et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020, 26, 681–687.
- Li, W.; Li, M.; Ou, G. COVID-19, cilia, and smell. FEBS J. 2020, 287, 3672–3676.
- Sternberg, A.; Naujokat, C. Structural features of coronavirus SARS-CoV-2 spike protein: Targets for vaccination. Life Sci. 2020, 257, 118056.
- Chen, M.; Pekosz, A.; Villano, J.S.; Shen, W.; Zhou, R.; Kulaga, H.; Li, Z.; Beck, S.E.; Witwer, K.W.; Mankowski, J.L.; et al. Evolution of nasal and olfactory infection characteristics of SARS-CoV-2 variants. bioRxiv 2022.
- Torabi, A.; Mohammadbagheri, E.; Akbari Dilmaghani, N.; Bayat, A.H.; Fathi, M.; Vakili, K.; Alizadeh, R.; Rezaeimirghaed, O.; Hajiesmaeili, M.; Ramezani, M.; et al. Proinflammatory Cytokines in the Olfactory Mucosa Result in COVID-19 Induced Anosmia. ACS Chem. Neurosci. 2020, 11, 1909–1913.
- von Bartheld, C.S.; Wang, L. Prevalence of Olfactory Dysfunction with the Omicron Variant of SARS-CoV-2: A Systematic Review and Meta-Analysis. Cells 2023, 12, 430.
- von Bartheld, C.S.; Hagen, M.M.; Butowt, R. The D614G Virus Mutation Enhances Anosmia in COVID-19 Patients: Evidence from a Systematic Review and Meta-analysis of Studies from South Asia. ACS Chem. Neurosci. 2021, 12, 3535–3549.
- Schreiner, T.; Allnoch, L.; Beythien, G.; Marek, K.; Becker, K.; Schaudien, D.; Stanelle-Bertram, S.; Schaumburg, B.; Mounogou Kouassi, N.; Beck, S.; et al. SARS-CoV-2 Infection Dysregulates Cilia and Basal Cell Homeostasis in the Respiratory Epithelium of Hamsters. Int. J. Mol. Sci. 2022, 23, 5124.
- Seehusen, F.; Clark, J.J.; Sharma, P.; Bentley, E.G.; Kirby, A.; Subramaniam, K.; Wunderlin-Giuliani, S.; Hughes, G.L.; Patterson, E.I.; Michael, B.D.; et al. Neuroinvasion and Neurotropism by SARS-CoV-2 Variants in the K18-hACE2 Mouse. Viruses 2022, 14, 1020.
- Zheng, J.; Wong, L.R.; Li, K.; Verma, A.K.; Ortiz, M.E.; Wohlford-Lenane, C.; Leidinger, M.R.; Knudson, C.M.; Meyerholz, D.K.; McCray, P.B., Jr.; et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 2021, 589, 603–607.
- Trobaugh, D.W.; Klimstra, W.B. MicroRNA Regulation of RNA Virus Replication and Pathogenesis. Trends Mol. Med. 2017, 23, 80–93.
- Hanna, J.; Hossain, G.S.; Kocerha, J. The Potential for microRNA Therapeutics and Clinical Research. Front. Genet. 2019, 10, 478.
- Osan, J.K.; DeMontigny, B.A.; Mehedi, M. Immunohistochemistry for protein detection in PFA-fixed paraffin-embedded SARS-CoV-2-infected COPD airway epithelium. STAR Protoc. 2021, 2, 100663.
- Davies, J.; Randeva, H.S.; Chatha, K.; Hall, M.; Spandidos, D.A.; Karteris, E.; Kyrou, I. Neuropilin-1 as a new potential SARS-CoV-2 infection mediator implicated in the neurologic features and central nervous system involvement of COVID-19. Mol. Med. Rep. 2020, 22, 4221–4226.
- Kang, Y.L.; Chou, Y.Y.; Rothlauf, P.W.; Liu, Z.; Soh, T.K.; Cureton, D.; Case, J.B.; Chen, R.E.; Diamond, M.S.; Whelan, S.P.J.; et al. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 20803–20813.
- Mayi, B.S.; Leibowitz, J.A.; Woods, A.T.; Ammon, K.A.; Liu, A.E.; Raja, A. The role of Neuropilin-1 in COVID-19. PLoS Pathog. 2021, 17, e1009153.
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860.
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Anton-Plagaro, C.; Shoemark, D.K.; Simon-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865.
More
Information
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
513
Revisions:
2 times
(View History)
Update Date:
18 Mar 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




