
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Maria-Anna Kyrgiafini | -- | 2174 | 2024-03-10 22:17:15 | | | |
| 2 | Lindsay Dong | Meta information modification | 2174 | 2024-03-11 03:11:34 | | |
Video Upload Options
Circular RNAs (circRNAs) are derived from regions that are transcribed but do not code for proteins. Instead, they form covalent closed-loop structures and play a crucial role in many biological processes. Studies show that they have an important role in cattle health, welfare and productive characteristics.
1. Introduction
2. Circular RNAs in Cattle
Cattle (Bos taurus) play a pivotal role in global agriculture, functioning as key economic keystones and nutritional reservoirs with far-reaching significance. As integral contributors to the meat and dairy industries, cattle have great economic importance. The mandate to optimize their productivity, health, and welfare converges at the intersection of economic sustainability and scientific progress, particularly in light of the many challenges faced by cattle farming, encompassing disease susceptibility [21], productivity oscillations, and environmental stressors [22][23]. The study of circRNAs can provide a better understanding of the intricate molecular mechanisms governing cattle physiology, presenting avenues for targeted interventions and innovative strategies to address the multifaceted challenges encountered by the cattle farming industry.
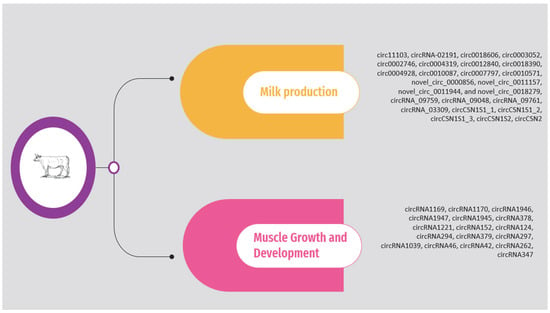
2.1. Milk Production
2.2. Muscle Growth and Development
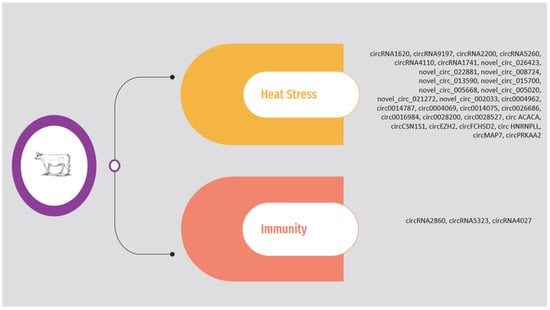
2.3. Immunity
2.4. Heat Stress
References
- Salmon, G.; MacLeod, M.; Claxton, J.; Ciamarra, U.P.; Robinson, T.; Duncan, A.; Peters, A. Exploring the landscape of livestock ‘Facts’. Glob. Food Secur. 2020, 25, 100329.
- Westhoek, H.; Lesschen, J.; Leip, A.; Rood, T.; Wagner, S.; De Marco, A.; Murphy-Brokern, D.; Palliere, C.; Howard, C.; Oenema, O.; et al. Nitrogen on the Table: The Influence of Food Choices on Nitrogen Emissions, Greenhouse Gas Emissions and Land Use in Europe; Centre for Ecology & Hydrology: Edinburgh, UK, 2015.
- Akinmoladun, O.F.; Muchenje, V.; Fon, F.N.; Mpendulo, C.T. Small Ruminants: Farmers’ Hope in a World Threatened by Water Scarcity. Animals 2019, 9, 456.
- Besbes, B.; Alary, V.; Baltenweck, I. Livestock Sector Investment and Policy Toolkit (LSIPT), Making Responsible Decisions; FAO: Rome, Italy, 2020.
- Cheng, M.; McCarl, B.; Fei, C. Climate Change and Livestock Production: A Literature Review. Atmosphere 2022, 13, 140.
- Daszkiewicz, T. Food Production in the Context of Global Developmental Challenges. Agriculture 2022, 12, 832.
- Jones, B.A.; Grace, D.; Kock, R.; Alonso, S.; Rushton, J.; Said, M.Y.; McKeever, D.; Mutua, F.; Young, J.; McDermott, J.; et al. Zoonosis Emergence Linked to Agricultural Intensification and Environmental Change. Proc. Natl. Acad. Sci. USA 2013, 110, 8399–8404.
- Miretti, S.; Lecchi, C.; Ceciliani, F.; Baratta, M. MicroRNAs as Biomarkers for Animal Health and Welfare in Livestock. Front. Vet. Sci. 2020, 7, 578193.
- Chen, H.; Shan, G. The Physiological Function of Long-Noncoding RNAs. Non-Coding RNA Res. 2020, 5, 178–184.
- Stefani, G.; Slack, F.J. Small Non-Coding RNAs in Animal Development. Nat. Rev. Mol. Cell Biol. 2008, 9, 219–230.
- Lagarrigue, S.; Lorthiois, M.; Degalez, F.; Gilot, D.; Derrien, T. LncRNAs in Domesticated Animals: From Dog to Livestock Species. Mamm. Genome 2022, 33, 248–270.
- Winter, E.; Cisilotto, J.; Silva, A.H.; Rosolen, D.; Fabichak, A.P.; Rode, M.P.; Creczynski-Pasa, T.B. MicroRNAs: Potential Biomarkers for Reproduction, Diagnosis, Prognosis, and Therapeutic in Domestic Animals. Res. Vet. Sci. 2022, 142, 117–132.
- Shaw, S.; Khurana, S.; Mukherjee, A.; Nayak, R.; Bose, S. Clinical Utility of Noncoding RNAs as Systemic Biomarkers in Animal Models. In Handbook of Animal Models and Its Uses in Cancer Research; Springer Nature: Singapore, 2023; pp. 1107–1123.
- Greene, J.; Baird, A.-M.; Brady, L.; Lim, M.; Gray, S.G.; McDermott, R.; Finn, S.P. Circular RNAs: Biogenesis, Function and Role in Human Diseases. Front. Mol. Biosci. 2017, 4, 38.
- Chen, L.-L.; Yang, L. Regulation of circRNA Biogenesis. RNA Biol. 2015, 12, 381–388.
- Chen, L.; Huang, C.; Wang, X.; Shan, G. Circular RNAs in Eukaryotic Cells. Curr. Genom. 2015, 16, 312–318.
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A New Star of Noncoding RNAs. Cancer Lett. 2015, 365, 141–148.
- Patop, I.L.; Wüst, S.; Kadener, S. Past, Present, and Future of Circ RNA s. EMBO J. 2019, 38, e100836.
- Guria, A.; Sharma, P.; Natesan, S.; Pandi, G. Circular RNAs—The Road Less Traveled. Front. Mol. Biosci. 2020, 6, 146.
- Quan, G.; Li, J. Circular RNAs: Biogenesis, Expression and Their Potential Roles in Reproduction. J. Ovarian Res. 2018, 11, 9.
- LeBlanc, S.J.; Lissemore, K.D.; Kelton, D.F.; Duffield, T.F.; Leslie, K.E. Major Advances in Disease Prevention in Dairy Cattle. J. Dairy Sci. 2006, 89, 1267–1279.
- Hume, D.A.; Whitelaw, C.B.A.; Archibald, A.L. The Future of Animal Production: Improving Productivity and Sustainability. J. Agric. Sci. 2011, 149, 9–16.
- Gauly, M.; Bollwein, H.; Breves, G.; Brügemann, K.; Dänicke, S.; Daş, G.; Demeler, J.; Hansen, H.; Isselstein, J.; König, S.; et al. Future Consequences and Challenges for Dairy Cow Production Systems Arising from Climate Change in Central Europe—A review. Animal 2013, 7, 843–859.
- Strucken, E.M.; Laurenson, Y.C.S.M.; Brockmann, G.A. Go with the Flow-Biology and Genetics of the Lactation Cycle. Front. Genet. 2015, 6, 118.
- FAO Dairy and Dairy Products. OECD-FAO Agricultural Outlook 2020–2029; FAO: Paris, France, 2020.
- Hanuš, O.; Samková, E.; Křížová, L.; Hasoňová, L.; Kala, R. Role of Fatty Acids in Milk Fat and the Influence of Selected Factors on Their Variability—A Review. Molecules 2018, 23, 1636.
- Zhang, C.; Wu, H.; Wang, Y.; Zhu, S.; Liu, J.; Fang, X.; Chen, H. Circular RNA of Cattle Casein Genes Are Highly Expressed in Bovine Mammary Gland. J. Dairy Sci. 2016, 99, 4750–4760.
- Chen, Z.; Lu, Q.; Liang, Y.; Cui, X.; Wang, X.; Mao, Y.; Yang, Z. Circ11103 Interacts with MiR-128/PPARGC1A to Regulate Milk Fat Metabolism in Dairy Cows. J. Agric. Food Chem. 2021, 69, 4490–4500.
- Liang, Y.; Gao, Q.; Wang, H.; Guo, M.; Arbab, A.A.I.; Nazar, M.; Li, M.; Yang, Z.; Karrow, N.A.; Mao, Y. Identification and Characterization of Circular RNAs in Mammary Tissue from Holstein Cows at Early Lactation and Non-Lactation. Biomolecules 2022, 12, 478.
- Feng, X.; Cai, Z.; Mu, T.; Yu, B.; Wang, Y.; Ma, R.; Liu, J.; Wang, C.; Zhang, J.; Gu, Y. CircRNA Screening and ceRNA Network Construction for Milk Fat Metabolism in Dairy Cows. Front. Vet. Sci. 2022, 9, 995629.
- González, N.; Marquès, M.; Nadal, M.; Domingo, J.L. Meat Consumption: Which Are the Current Global Risks? A Review of Recent (2010–2020) Evidences. Food Res. Int. 2020, 137, 109341.
- Milford, A.B.; Le Mouël, C.; Bodirsky, B.L.; Rolinski, S. Drivers of Meat Consumption. Appetite 2019, 141, 104313.
- Oksbjerg, N.; Therkildsen, M. Myogenesis and Muscle Growth and Meat Quality. In New Aspects of Meat Quality; Woodhead Publishing: Sawston, UK, 2017; pp. 33–62.
- Wei, X.; Li, H.; Yang, J.; Hao, D.; Dong, D.; Huang, Y.; Lan, X.; Plath, M.; Lei, C.; Lin, F.; et al. Circular RNA Profiling Reveals an Abundant circLMO7 That Regulates Myoblasts Differentiation and Survival by Sponging miR-378a-3p. Cell Death Dis. 2017, 8, e3153.
- Ingvartsen, K.L.; Moyes, K. Nutrition, Immune Function and Health of Dairy Cattle. Animal 2013, 7, 112–122.
- Vlasova, A.N.; Saif, L.J. Bovine Immunology: Implications for Dairy Cattle. Front. Immunol. 2021, 12, 643206.
- Bai, Z.; Wu, Y.; Cai, W.; Zheng, Y.; Hui, T.; Yue, C.; Sun, J.; Wang, Y.; Xu, Z.; Wang, Z. High-Throughput Analysis of CircRNA in Cows with Naturally Infected Staphylococcus Aureus Mammary Gland. Anim. Biotechnol. 2023, 34, 4236–4246.
- Polsky, L.; von Keyserlingk, M.A. Invited Review: Effects of Heat Stress on Dairy Cattle Welfare. J. Dairy Sci. 2017, 100, 8645–8657.
- Fernandez-Novo, A.; Pérez-Garnelo, S.S.; Villagrá, A.; Pérez-Villalobos, N.; Astiz, S. The Effect of Stress on Reproduction and Reproductive Technologies in Beef Cattle—A Review. Animals 2020, 10, 2096.
- Herbut, P.; Angrecka, S.; Godyń, D.; Hoffmann, G. The Physiological and Productivity Effects of Heat Stress in Cattle—A Review. Ann. Anim. Sci. 2019, 19, 579–593.
- Zhang, C.; Wang, S.; Hu, L.; Fang, H.; Chen, G.; Ma, X.; Yu, Y.; Wang, Y.; Xu, Q. Analysis of CircRNA Expression in Peripheral Blood of Holstein Cows in Response to Heat Stress. Int. J. Mol. Sci. 2023, 24, 10150.
- Zeng, H.; Xia, H.; Wang, X.; Wang, Y.; Fang, J.; Li, S.; Zhai, Y.; Han, Z. Comprehensive Profiling of CeRNA (CircRNA-MiRNA-MRNA) Networks in Hypothalamic-Pituitary-Mammary Gland Axis of Dairy Cows under Heat Stress. Int. J. Mol. Sci. 2023, 24, 888.




