
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Stéphane Martin | -- | 3940 | 2024-03-07 09:57:59 | | | |
| 2 | Stéphane Martin | Meta information modification | 3940 | 2024-03-07 10:01:27 | | | | |
| 3 | Lindsay Dong | Meta information modification | 3940 | 2024-03-08 02:19:35 | | | | |
| 4 | Lindsay Dong | Meta information modification | 3940 | 2024-03-08 06:16:08 | | |
Video Upload Options
Neurotransmission occurs within highly specialized compartments forming the active synapse where the complex organization and dynamics of the interactions are tightly orchestrated both in time and space. Post-translational modifications (PTMs) are central to these spatiotemporal regulations to ensure an efficient synaptic transmission. SUMOylation is a dynamic PTM that modulates the interactions between proteins and consequently regulates the conformation, the distribution and the trafficking of the SUMO-target proteins. SUMOylation plays a crucial role in synapse formation and stabilization, as well as in the regulation of synaptic transmission and plasticity.
1. Introduction
2. The SUMOylation/deSUMOylation Enzymatic Pathway at Synapses
2.1. SUMO Paralogs and the Associated Enzymatic SUMOylation/deSUMOylation Cascade
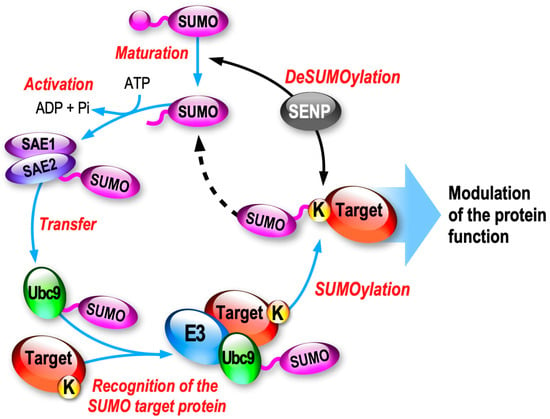
2.2. Synaptic Localization of the SUMOylation/deSUMOylation Machinery
2.3. Synaptic Regulation of the SUMO Pathway
An additional level of activity-dependent regulation of the SUMO/deSUMOylation pathway occurs directly at the synapse and relies on the activation of type 1 metabotropic glutamate mGlu1 and mGluR5 receptors [1[9][23][24]. A short activation of mGlu5 receptors triggers a Protein Kinase C (PKC) phosphorylation-dependent trapping of the SUMO-conjugating enzyme Ubc9 at post-synaptic sites, leading to an increased SUMOylation and to the modulation of neuronal excitability [1[9]. The sustained activation of mGlu5R then leads to a decrease in the exit rate of the deSUMOylation enzyme SENP1 from dendritic spines resulting in the post-synaptic accumulation of SENP1 to bring SUMOylation back to initial levels [23]. Blocking the activation of mGlu1R enhances the mGlu5R-induced post-synaptic accumulation of SENP1 indicating that the SUMOylation/deSUMOylation balance is bidirectionally regulated by type I mGluRs to control the levels of synaptic SUMOylation [24]. Altogether, these findings revealed that type I mGlu1/5 receptors are central to dynamically maintain the homeostasis of SUMOylation at the mammalian synapse but also indicate that the relative amount of mGlu1 and mGlu5 receptors at individual synapses will drive the diversity of the post-synaptic SUMOylation responses.
3. SUMOylation in Neurite Growth, Synapse Formation, Elimination and Maturation
3.1. SUMOylation of Transcription Factors in Neurite Growth and Branching
3.1.1. MEF2 SUMOylation
3.1.2. FOXP2 SUMOylation
3.1.3. MeCP2 SUMOylation
3.1.4. ZBTB20 SUMOylation
3.2. Extranuclear SUMOylation in Synapse Formation and Maturation
3.2.1. CASK SUMOylation
3.2.2. Local Protein Synthesis and Dendritic SUMOylation
CPEB3 SUMOylation
The RNA-binding CPEB3 belongs to a family of four proteins in vertebrates and is involved in the regulation of local translation in the brain, and consequently in synaptic plasticity and memory formation. The Kandel lab first reported the SUMO2-ylated CPEB3 in hippocampal neurons both in vitro and in vivo [39]. They showed that the SUMOylation of CPEB3 regulates its oligomerization. In a basal state, CPEB3 is SUMOylated and acts as a local translational repressor. CPEB3 is rapidly deSUMOylated upon neuronal activation, leading to its aggregation and the translation of target mRNAs, including the sumo2 mRNA [39].
FMRP SUMOylation
The RNA-binding protein FMRP participates in the formation and organization of membraneless RNA-containing structures in dendrites and axons to transport and locally regulate the translation of several mRNAs essential for synaptic formation and plasticity in a mGlu5 receptor-dependent manner [40][41][42][43]. Loss of FMRP expression or function leads to the most common inherited cause of intellectual disability and autism called Fragile X Syndrome (FXS; [42][43]). At the cellular level, the absence of FMRP leads to a pathological hyperabundance of immature spines in FXS patients and mouse models of the disease [44][45]. The consequences of these synaptic defects are synaptic transmission and plasticity alterations leading to socio-cognitive impairments.
3.3. SUMOylation and Microtubules (MTs)
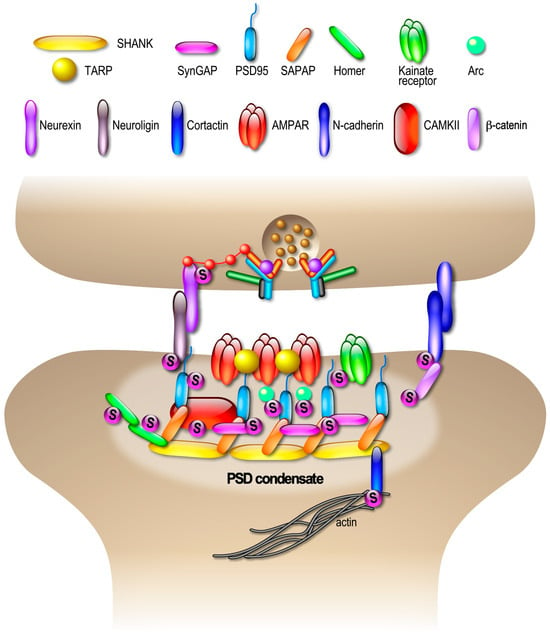
4. SUMOylation, Biomolecular Condensates and Compartmentalization of the Synapse
4.1. En Masse SUMOylation at Synapses?
4.2. SUMOylation and LLPS
4.3. SUMOylation and Compartmentalization of Pre- and Post-Synaptic Sites
References
- Matunis, M.J.; Coutavas, E.; Blobel, G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996, 135, 1457–1470.
- Mahajan, R.; Delphin, C.; Guan, T.; Gerace, L.; Melchior, F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 1997, 88, 97–107.
- Flotho, A.; Melchior, F. Sumoylation: A regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013, 82, 357–385.
- Celen, A.B.; Sahin, U. Sumoylation on its 25th anniversary: Mechanisms, pathology, and emerging concepts. FEBS J. 2020, 287, 3110–3140.
- Gwizdek, C.; Casse, F.; Martin, S. Protein sumoylation in brain development, neuronal morphology and spinogenesis. Neuromol. Med. 2013, 15, 677–691.
- Schorova, L.; Martin, S. Sumoylation in Synaptic Function and Dysfunction. Front. Synaptic Neurosci. 2016, 8, 9.
- Henley, J.M.; Carmichael, R.E.; Wilkinson, K.A. Extranuclear SUMOylation in Neurons. Trends Neurosci. 2018, 41, 198–210.
- Folci, A.; Mirabella, F.; Fossati, M. Ubiquitin and Ubiquitin-Like Proteins in the Critical Equilibrium between Synapse Physiology and Intellectual Disability. eNeuro 2020, 7, 1–25.
- Henley, J.M.; Seager, R.; Nakamura, Y.; Talandyte, K.; Nair, J.; Wilkinson, K.A. SUMOylation of synaptic and synapse-associated proteins: An update. J. Neurochem. 2021, 156, 145–161.
- Colnaghi, L.; Russo, L.; Natale, C.; Restelli, E.; Cagnotto, A.; Salmona, M.; Chiesa, R.; Fioriti, L. Super Resolution Microscopy of SUMO Proteins in Neurons. Front. Cell. Neurosci. 2019, 13, 486.
- Colnaghi, L.; Rondelli, D.; Muzi-Falconi, M.; Sertic, S. Tau and DNA Damage in Neurodegeneration. Brain Sci. 2020, 10, 946.
- Conz, A.; Musi, C.A.; Russo, L.; Borsello, T.; Colnaghi, L. Super-resolution study of PIAS SUMO E3-ligases in hippocampal and cortical neurons. Eur. J. Histochem. 2021, 65, 3241.
- Martin, S.; Nishimune, A.; Mellor, J.R.; Henley, J.M. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature 2007, 447, 321–325.
- Feligioni, M.; Nishimune, A.; Henley, J.M. Protein SUMOylation modulates calcium influx and glutamate release from presynaptic terminals. Eur. J. Neurosci. 2009, 29, 1348–1356.
- Loriol, C.; Parisot, J.; Poupon, G.; Gwizdek, C.; Martin, S. Developmental regulation and spatiotemporal redistribution of the sumoylation machinery in the rat central nervous system. PLoS ONE 2012, 7, e33757.
- Loriol, C.; Khayachi, A.; Poupon, G.; Gwizdek, C.; Martin, S. Activity-dependent regulation of the sumoylation machinery in rat hippocampal neurons. Biol. Cell 2013, 105, 30–45.
- Loriol, C.; Casse, F.; Khayachi, A.; Poupon, G.; Chafai, M.; Deval, E.; Gwizdek, C.; Martin, S. mGlu5 receptors regulate synaptic sumoylation via a transient PKC-dependent diffusional trapping of Ubc9 into spines. Nat. Commun. 2014, 5, 5113.
- Hasegawa, Y.; Yoshida, D.; Nakamura, Y.; Sakakibara, S. Spatiotemporal distribution of SUMOylation components during mouse brain development. J. Comp. Neurol. 2014, 522, 3020–3036.
- Josa-Prado, F.; Luo, J.; Rubin, P.; Henley, J.M.; Wilkinson, K.A. Developmental profiles of SUMOylation pathway proteins in rat cerebrum and cerebellum. PLoS ONE 2019, 14, e0212857.
- Guo, C.; Hildick, K.L.; Luo, J.; Dearden, L.; Wilkinson, K.A.; Henley, J.M. SENP3-mediated deSUMOylation of dynamin-related protein 1 promotes cell death following ischaemia. EMBO J. 2013, 32, 1514–1528.
- Waters, E.; Wilkinson, K.A.; Harding, A.L.; Carmichael, R.E.; Robinson, D.; Colley, H.E.; Guo, C. The SUMO protease SENP3 regulates mitochondrial autophagy mediated by Fis1. EMBO Rep. 2022, 23, e48754.
- Jaafari, N.; Konopacki, F.A.; Owen, T.F.; Kantamneni, S.; Rubin, P.; Craig, T.J.; Wilkinson, K.A.; Henley, J.M. SUMOylation is required for glycine-induced increases in AMPA receptor surface expression (ChemLTP) in hippocampal neurons. PLoS ONE 2013, 8, e52345.
- Lyons, G.E.; Micales, B.K.; Schwarz, J.; Martin, J.F.; Olson, E.N. Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J. Neurosci. 1995, 15, 5727–5738.
- Flavell, S.W.; Greenberg, M.E. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu. Rev. Neurosci. 2008, 31, 563–590.
- Shalizi, A.; Gaudilliere, B.; Yuan, Z.; Stegmuller, J.; Shirogane, T.; Ge, Q.; Tan, Y.; Schulman, B.; Harper, J.W.; Bonni, A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science 2006, 311, 1012–1017.
- Lu, H.; Liu, B.; You, S.; Chen, L.; Dongmei, Q.; Gu, M.; Lu, Y.; Chen, Y.; Zhang, F.; Yu, B. SENP2 regulates MEF2A de-SUMOylation in an activity dependent manner. Mol. Biol. Rep. 2013, 40, 2485–2490.
- Meredith, L.J.; Wang, C.M.; Nascimento, L.; Liu, R.; Wang, L.; Yang, W.H. The Key Regulator for Language and Speech Development, FOXP2, is a Novel Substrate for SUMOylation. J. Cell. Biochem. 2016, 117, 426–438.
- Estruch, S.B.; Graham, S.A.; Deriziotis, P.; Fisher, S.E. The language-related transcription factor FOXP2 is post-translationally modified with small ubiquitin-like modifiers. Sci. Rep. 2016, 6, 20911.
- Usui, N.; Co, M.; Harper, M.; Rieger, M.A.; Dougherty, J.D.; Konopka, G. Sumoylation of FOXP2 Regulates Motor Function and Vocal Communication Through Purkinje Cell Development. Biol. Psychiatry 2017, 81, 220–230.
- Cheng, J.; Huang, M.; Zhu, Y.; Xin, Y.J.; Zhao, Y.K.; Huang, J.; Yu, J.X.; Zhou, W.H.; Qiu, Z. SUMOylation of MeCP2 is essential for transcriptional repression and hippocampal synapse development. J. Neurochem. 2014, 128, 798–806.
- Zhang, W.; Mi, J.; Li, N.; Sui, L.; Wan, T.; Zhang, J.; Chen, T.; Cao, X. Identification and characterization of DPZF, a novel human BTB/POZ zinc finger protein sharing homology to BCL-6. Biochem. Biophys. Res. Commun. 2001, 282, 1067–1073.
- Gilman, S.R.; Iossifov, I.; Levy, D.; Ronemus, M.; Wigler, M.; Vitkup, D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron 2011, 70, 898–907.
- Jones, K.A.; Luo, Y.; Dukes-Rimsky, L.; Srivastava, D.P.; Koul-Tewari, R.; Russell, T.A.; Shapiro, L.P.; Srivastava, A.K.; Penzes, P. Neurodevelopmental disorder-associated ZBTB20 gene variants affect dendritic and synaptic structure. PLoS ONE 2018, 13, e0203760.
- Ripamonti, S.; Shomroni, O.; Rhee, J.S.; Chowdhury, K.; Jahn, O.; Hellmann, K.P.; Bonn, S.; Brose, N.; Tirard, M. SUMOylation controls the neurodevelopmental function of the transcription factor Zbtb20. J. Neurochem. 2020, 154, 647–661.
- Biederer, T.; Sudhof, T.C. CASK and protein 4.1 support F-actin nucleation on neurexins. J. Biol. Chem. 2001, 276, 47869–47876.
- Chao, H.W.; Hong, C.J.; Huang, T.N.; Lin, Y.L.; Hsueh, Y.P. SUMOylation of the MAGUK protein CASK regulates dendritic spinogenesis. J. Cell Biol. 2008, 182, 141–155.
- Drisaldi, B.; Colnaghi, L.; Fioriti, L.; Rao, N.; Myers, C.; Snyder, A.M.; Metzger, D.J.; Tarasoff, J.; Konstantinov, E.; Fraser, P.E.; et al. SUMOylation Is an Inhibitory Constraint that Regulates the Prion-like Aggregation and Activity of CPEB3. Cell Rep. 2015, 11, 1694–1702.
- Huber, K.M.; Roder, J.C.; Bear, M.F. Chemical induction of mGluR5- and protein synthesis--dependent long-term depression in hippocampal area CA1. J. Neurophysiol. 2001, 86, 321–325.
- Bear, M.F.; Huber, K.M.; Warren, S.T. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004, 27, 370–377.
- Bassell, G.J. Fragile balance: RNA editing tunes the synapse. Nat. Neurosci. 2011, 14, 1492–1494.
- Darnell, J.C.; Van Driesche, S.J.; Zhang, C.; Hung, K.Y.; Mele, A.; Fraser, C.E.; Stone, E.F.; Chen, C.; Fak, J.J.; Chi, S.W.; et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 2011, 146, 247–261.
- The Dutch-Belgian Fragile X Consortium; Bakker, C.E.; Verheij, C.; Willemsen, R.; van der Helm, R.; Oerlemans, F.; Vermey, M.; Bygrave, A.; Hoogeveen, A.; Oostra, B.A.; et al. Fmr1 knockout mice: A model to study fragile X mental retardation. The Dutch-Belgian Fragile X Consortium. Cell 1994, 78, 23–33.
- Mientjes, E.J.; Nieuwenhuizen, I.; Kirkpatrick, L.; Zu, T.; Hoogeveen-Westerveld, M.; Severijnen, L.; Rife, M.; Willemsen, R.; Nelson, D.L.; Oostra, B.A. The generation of a conditional Fmr1 knock out mouse model to study Fmrp function in vivo. Neurobiol. Dis. 2006, 21, 549–555.
- Baas, P.W.; Rao, A.N.; Matamoros, A.J.; Leo, L. Stability properties of neuronal microtubules. Cytoskeleton 2016, 73, 442–460.
- Gu, J.; Firestein, B.L.; Zheng, J.Q. Microtubules in dendritic spine development. J. Neurosci. 2008, 28, 12120–12124.
- Mitchison, T.; Kirschner, M. Dynamic instability of microtubule growth. Nature 1984, 312, 237–242.
- Li, S.; Liang, Y.; Zou, J.; Cai, Z.; Yang, H.; Yang, J.; Zhang, Y.; Lin, H.; Zhang, G.; Tan, M. SUMOylation of microtubule-cleaving enzyme KATNA1 promotes microtubule severing and neurite outgrowth. J. Biol. Chem. 2022, 298, 102292.
- McNally, F.J.; Vale, R.D. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell 1993, 75, 419–429.
- Chen, K.; Ye, Y.; Ji, Z.; Tan, M.; Li, S.; Zhang, J.; Guo, G.; Lin, H. Katanin p60 promotes neurite growth and collateral formation in the hippocampus. Int. J. Clin. Exp. Med. 2014, 7, 2463–2470.
- Shin, S.C.; Im, S.K.; Jang, E.H.; Jin, K.S.; Hur, E.M.; Kim, E.E. Structural and Molecular Basis for Katanin-Mediated Severing of Glutamylated Microtubules. Cell Rep. 2019, 26, 1357–1367.e5.
- Ji, Z.S.; Liu, Q.L.; Zhang, J.F.; Yang, Y.H.; Li, J.; Zhang, G.W.; Tan, M.H.; Lin, H.S.; Guo, G.Q. SUMOylation of spastin promotes the internalization of GluA1 and regulates dendritic spine morphology by targeting microtubule dynamics. Neurobiol. Dis. 2020, 146, 105133.
- Yu, W.; Qiang, L.; Solowska, J.M.; Karabay, A.; Korulu, S.; Baas, P.W. The microtubule-severing proteins spastin and katanin participate differently in the formation of axonal branches. Mol. Biol. Cell 2008, 19, 1485–1498.
- Vertegaal, A.C.O. Signalling mechanisms and cellular functions of SUMO. Nat. Rev. Mol. Cell Biol. 2022, 23, 715–731.
- Gasser, S.M.; Stutz, F. SUMO in the regulation of DNA repair and transcription at nuclear pores. FEBS Lett. 2023, 597, 2833–2850.
- Esteras, M.; Liu, I.C.; Snijders, A.P.; Jarmuz, A.; Aragon, L. Identification of SUMO conjugation sites in the budding yeast proteome. Microb. Cell 2017, 4, 331–341.
- Pronot, M.; Kieffer, F.; Gay, A.S.; Debayle, D.; Forquet, R.; Poupon, G.; Schorova, L.; Martin, S.; Gwizdek, C. Proteomic Identification of an Endogenous Synaptic SUMOylome in the Developing Rat Brain. Front. Mol. Neurosci. 2021, 14, 780535.
- Psakhye, I.; Jentsch, S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 2012, 151, 807–820.
- Cheng, X.; Yang, W.; Lin, W.; Mei, F. Paradoxes of Cellular SUMOylation Regulation: A Role of Biomolecular Condensates? Pharmacol. Rev. 2023, 75, 979–1006.
- Keiten-Schmitz, J.; Roder, L.; Hornstein, E.; Muller-McNicoll, M.; Muller, S. SUMO: Glue or Solvent for Phase-Separated Ribonucleoprotein Complexes and Molecular Condensates? Front. Mol. Biosci. 2021, 8, 673038.
- Bratek-Skicki, A.; Pancsa, R.; Meszaros, B.; Van Lindt, J.; Tompa, P. A guide to regulation of the formation of biomolecular condensates. FEBS J. 2020, 287, 1924–1935.
- Yau, T.Y.; Sander, W.; Eidson, C.; Courey, A.J. SUMO Interacting Motifs: Structure and Function. Cells 2021, 10, 2825.
- Lascorz, J.; Codina-Fabra, J.; Reverter, D.; Torres-Rosell, J. SUMO-SIM interactions: From structure to biological functions. Semin. Cell Dev. Biol. 2022, 132, 193–202.
- Rodriguez, M.S.; Dargemont, C.; Hay, R.T. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001, 276, 12654–12659.
- Rizo, J. Mechanism of neurotransmitter release coming into focus. Protein Sci. 2018, 27, 1364–1391.
- Benson, M.; Iniguez-Lluhi, J.A.; Martens, J. Sumo Modification of Ion Channels. Adv. Exp. Med. Biol. 2017, 963, 127–141.
- Tang, L.T.; Craig, T.J.; Henley, J.M. SUMOylation of synapsin Ia maintains synaptic vesicle availability and is reduced in an autism mutation. Nat. Commun. 2015, 6, 7728.
- Sansevrino, R.; Hoffmann, C.; Milovanovic, D. Condensate biology of synaptic vesicle clusters. Trends Neurosci. 2023, 46, 293–306.
- Rousseaux, M.W.C.; Vazquez-Velez, G.E.; Al-Ramahi, I.; Jeong, H.H.; Bajic, A.; Revelli, J.P.; Ye, H.; Phan, E.T.; Deger, J.M.; Perez, A.M.; et al. A Druggable Genome Screen Identifies Modifiers of α-Synuclein Levels via a Tiered Cross-Species Validation Approach. J. Neurosci. 2018, 38, 9286–9301.
- Krumova, P.; Meulmeester, E.; Garrido, M.; Tirard, M.; Hsiao, H.H.; Bossis, G.; Urlaub, H.; Zweckstetter, M.; Kugler, S.; Melchior, F.; et al. Sumoylation inhibits α-synuclein aggregation and toxicity. J. Cell Biol. 2011, 194, 49–60.
- Leenders, A.G.; Sheng, Z.H. Modulation of neurotransmitter release by the second messenger-activated protein kinases: Implications for presynaptic plasticity. Pharmacol. Ther. 2005, 105, 69–84.
- Coppola, T.; Magnin-Luthi, S.; Perret-Menoud, V.; Gattesco, S.; Schiavo, G.; Regazzi, R. Direct interaction of the Rab3 effector RIM with Ca2+ channels, SNAP-25, and synaptotagmin. J. Biol. Chem. 2001, 276, 32756–32762.
- Girach, F.; Craig, T.J.; Rocca, D.L.; Henley, J.M. RIM1α SUMOylation is required for fast synaptic vesicle exocytosis. Cell Rep. 2013, 5, 1294–1301.
- Matsuzaki, S.; Lee, L.; Knock, E.; Srikumar, T.; Sakurai, M.; Hazrati, L.N.; Katayama, T.; Staniszewski, A.; Raught, B.; Arancio, O.; et al. SUMO1 Affects Synaptic Function, Spine Density and Memory. Sci. Rep. 2015, 5, 10730.
- Craig, T.J.; Anderson, D.; Evans, A.J.; Girach, F.; Henley, J.M. SUMOylation of Syntaxin1A regulates presynaptic endocytosis. Sci. Rep. 2015, 5, 17669.
- Choi, J.H.; Park, J.Y.; Park, S.P.; Lee, H.; Han, S.; Park, K.H.; Suh, Y.H. Regulation of mGluR7 trafficking by SUMOylation in neurons. Neuropharmacology 2016, 102, 229–235.
- Zeng, M.; Shang, Y.; Araki, Y.; Guo, T.; Huganir, R.L.; Zhang, M. Phase Transition in Postsynaptic Densities Underlies Formation of Synaptic Complexes and Synaptic Plasticity. Cell 2016, 166, 1163–1175.e12.
- Zeng, M.; Diaz-Alonso, J.; Ye, F.; Chen, X.; Xu, J.; Ji, Z.; Nicoll, R.A.; Zhang, M. Phase Separation-Mediated TARP/MAGUK Complex Condensation and AMPA Receptor Synaptic Transmission. Neuron 2019, 104, 529–543.e6.
- Wu, X.; Hu, S.; Kang, X.; Wang, C. Synaptotagmins: Beyond Presynaptic Neurotransmitter Release. Neuroscientist 2020, 26, 9–15.
- Nicoll, R.A.; Tomita, S.; Bredt, D.S. Auxiliary subunits assist AMPA-type glutamate receptors. Science 2006, 311, 1253–1256.
- Araki, Y.; Zeng, M.; Zhang, M.; Huganir, R.L. Rapid dispersion of SynGAP from synaptic spines triggers AMPA receptor insertion and spine enlargement during LTP. Neuron 2015, 85, 173–189.
- Chamberlain, S.E.; Gonzalez-Gonzalez, I.M.; Wilkinson, K.A.; Konopacki, F.A.; Kantamneni, S.; Henley, J.M.; Mellor, J.R. SUMOylation and phosphorylation of GluK2 regulate kainate receptor trafficking and synaptic plasticity. Nat. Neurosci. 2012, 15, 845–852.
- Konopacki, F.A.; Jaafari, N.; Rocca, D.L.; Wilkinson, K.A.; Chamberlain, S.; Rubin, P.; Kantamneni, S.; Mellor, J.R.; Henley, J.M. Agonist-induced PKC phosphorylation regulates GluK2 SUMOylation and kainate receptor endocytosis. Proc. Natl. Acad. Sci. USA 2011, 108, 19772–19777.
- Craig, T.J.; Henley, J.M. SUMOylation, Arc and the regulation homeostatic synaptic scaling: Implications in health and disease. Commun. Integr. Biol. 2012, 5, 634–636.
- Nair, R.R.; Patil, S.; Tiron, A.; Kanhema, T.; Panja, D.; Schiro, L.; Parobczak, K.; Wilczynski, G.; Bramham, C.R. Dynamic Arc SUMOylation and Selective Interaction with F-Actin-Binding Protein Drebrin A in LTP Consolidation In Vivo. Front. Synaptic Neurosci. 2017, 9, 8.




