Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Cinzia Calvio | -- | 3327 | 2024-03-06 11:25:03 | | | |
| 2 | Camila Xu | Meta information modification | 3327 | 2024-03-07 02:10:36 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Alloun, W.; Calvio, C. Recovering Bioactive Compounds from Plant Waste. Encyclopedia. Available online: https://encyclopedia.pub/entry/55915 (accessed on 08 February 2026).
Alloun W, Calvio C. Recovering Bioactive Compounds from Plant Waste. Encyclopedia. Available at: https://encyclopedia.pub/entry/55915. Accessed February 08, 2026.
Alloun, Wiem, Cinzia Calvio. "Recovering Bioactive Compounds from Plant Waste" Encyclopedia, https://encyclopedia.pub/entry/55915 (accessed February 08, 2026).
Alloun, W., & Calvio, C. (2024, March 06). Recovering Bioactive Compounds from Plant Waste. In Encyclopedia. https://encyclopedia.pub/entry/55915
Alloun, Wiem and Cinzia Calvio. "Recovering Bioactive Compounds from Plant Waste." Encyclopedia. Web. 06 March, 2024.
Copy Citation
Agro-industrial wastes are suitable as cost-effective sources of various health-promoting molecules at significant concentrations. lnvestigating new methods for converting them into high-value-added compounds is crucial for the sustainable development goals.
artificial intelligence
bioactive compounds
bioprocess digitization
Enzyme-assisted extraction
1. Introduction
The agrifood industry generates substantial volumes of waste across various food processing stages [1]. These residues often exhibit slow degradability due to their composition, leading to accumulation and adverse environmental impacts [2]. Plant wastes, however, are still rich in organic acids, minerals, vitamins, and secondary metabolites with beneficial effects on human health, and investigating new methods for converting them into high-value-added compounds is crucial for the sustainable development goals.
Various industrial sectors, such as food, nutraceuticals, pharmaceuticals, cosmetics, and agriculture, show a rising interest in new plant-originated active compounds, encompassing fibers, lipids, carbohydrates, peptides, secondary metabolites such as carotenoids, phenolics, and other multifunctional and bioactive components, as ingredients in different products [3].
Advanced approaches for the extraction of bioactive compounds, among which biological processes, can substitute for conventional chemical and solvent-based processes, and when the extraction parameters are optimized, biological extraction procedures can guarantee efficiency in the recovery and quality of the final products, while ensuring a reduced environmental impact [4][5]. Biological strategies include enzymatic extraction and microbial fermentation; these approaches provide numerous advantages over conventional methods in that they are more selective and have a milder impact on the bioactive molecules, generating high-quality extracts with pronounced bioactivity, maximal titer, and minimal toxicity [6].
Enzyme-assisted extraction (EAE) strategies ensure a controlled and effective process using enzymes to free compounds from the plant matrix [7]. Microbial fermentation strategies involve various microorganisms responsible for waste transformation into valuable resources such as phenolics, oligosaccharides, flavonoids, phytosterols, and others [8]. In addition to optimizing resource use, these biological approaches are compliant with the increasing focus on ecologically responsible and sustainable practices. Nonetheless, the efficiency of biological processes, such as enzymatic extraction and microbial fermentation, requires extensive trial-and-error before performances can overtake those of traditional harsh methodologies. This is due to the complex structure of the vegetable matrices, that varies in dependence of the chemical structure and physical properties of the substrate. Therefore, maximal performances of biological methods require the optimization of all parameters that can significantly affect enzyme activity or microbial fermentation, including the selection of the suitable biological pre-treatment and treatment and, for each of them, the tuning of the process conditions, i.e., temperature, pH, substrate concentration, and ionic strength reaction time.
Artificial intelligence (AI) and machine learning tools (ML) are emerging as pivotal iterative tools in digitizing and optimizing several interlinked processes. However, their application to the recovery of functional compounds from plant waste is in its infancy [9]. AI, combined with ML, can assist in identifying the optimal conditions for extraction processes by analyzing the complex data derived from substrate types, enzyme/microbe characteristics, and the variable treatment parameter’s options [10]. The AI-guided optimization of bioprocesses for the recovery of functional compounds from plant wastes leverages on the reliability of biological processes to maximize compound yield and titer while minimizing resource wastage and environmental impacts. These approaches enable the real-time monitoring and adjustment of extraction conditions, and the precise modelling and simulation of extraction procedures, enhancing understanding and efficiency.
2. Plant Waste as a Source of Functional Compounds
Agro-industrial waste contains various bioactive compounds, many of which are relevant for use in the food, cosmetics, and pharmaceutical industries and in plant protection products. These substances include bioactive peptides, phenolic compounds, polysaccharides, and other molecules possessing unique biological and technological qualities, as summarized in Table 1.
Table 1. Bioactive compounds, their sources, and their key properties and functions.
| Compound | Source | Properties and Functions | References |
|---|---|---|---|
| Bioactive Peptides | Cakes, meals, and plant by-products | Protein fragments (<20 amino acid residues) with diverse impacts on body functions. Antioxidant, antihypertensive, anti-inflammatory activities, and immune-modulating functions. When applied directly to food, they mitigate oxidation reactions, resulting in a safer alternative to synthetic antioxidants. |
Velliquette et al. [11] Hemker et al. [12] |
| Phenolic Compounds | Cereal bran, fruit and vegetable waste, complex carbohydrates | Antioxidant, antihypertensive, antimicrobial, and anti-carcinogenic effects. Widely used in the food industry to control lipid oxidation and microbial growth. Used in pharmaceutical and cosmetic industries, including mouthwashes, eye creams, and herbal cosmetics. They enhance the shelf-life of food products. |
Huang et al. [13] |
| Carbohydrates: Lignocellulose, β-glucans |
Starch, oat bran, and other cereal waste | Vital energy sources. Starch has widespread industrial applications. Lignocellulose (cellulose, hemicellulose, and lignin) can be converted into high-value products, contributing to waste reduction. β-Glucans, found in cereal waste, have scientifically proven health benefits with cholesterol-lowering and immune-modulating properties. |
Lovegrove et al. [14] Fortunati et al. [15] Tosh et al. [16] |
| Lycopene (Carotenoids) |
Tomato and by-products (skin and seeds) Carrots |
Natural pigment and antioxidant, suitable for food coloration and cellular protection. It prevents cellular components’ degradation, including DNA. β-Carotenoids are antioxidants and anti-inflammatory. |
Anarjan and Jouyban [17] Caseiro et al. [18] |
| Poly-unsaturated fatty acids (PUFAs): omega-3 and omega-6 | Vegetable oils, nuts, and their by-products | Anti-inflammatory agents. | Dave and Routray [19] |
In the food industry, these bioactive compounds can be directly incorporated into conventional food, improving its nutritional, sensory, and technological aspects. These improvements include gelatinization, foaming, emulsion stability, and the capacity to hold on to water and oil [20]. In addition, they might serve to create or modify films for use as intelligent and bioactive food packaging [21][22]. Moreover, these compounds can be used in designing functional foods, nutraceuticals, or food supplements, that can be used for the prevention and treatment of several diseases. Overall, bioactive compounds are associated with immune system protection, anti-inflammatory properties, decreased cell oxidative damage, and a decreased risk of developing chronic diseases (Figure 1) [23].
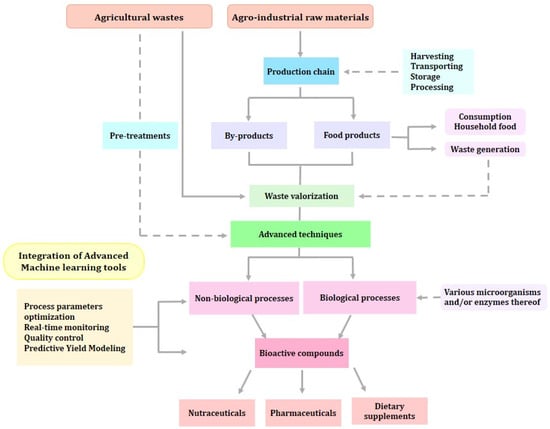
Figure 1. Overall process of plant waste valorization and recovery of functional compounds.
3. Conventional vs. Advanced Methods
The extraction of functional and nutraceutical compounds from plant waste can be achieved using various methods, depending on the specific compounds and source materials. These approaches consist of conventional (traditional) and advanced (sustainable) extraction methods (Table 2 and Figure 2).
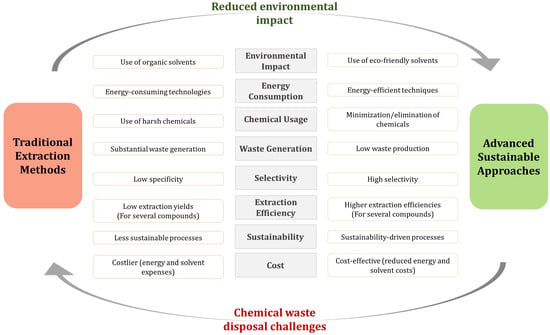
Figure 2. Differences between conventional and sustainable bioactive compound recovery methods.
Table 2. Conventional vs. advanced extraction methods of target bioactive metabolites from agro-industrial waste.
| Category | Traditional Approaches | Advanced Approaches | Target Bioactive Compounds | References |
|---|---|---|---|---|
| Solvent Extraction | Maceration | Supercritical CO2 Extraction | Phenolic compounds from grape pomace (resveratrol) | Carrasco-Sandoval et al. [24] |
| Soxhlet Extraction | Subcritical Water Extraction using pressurized hot water | Flavonoids from citrus peels (hesperidin) | Carrasco-Sandoval et al. [24] | |
| Hydro-distillation by steam | Ionic Liquid Extraction | Volatile compounds such as essential oils from aromatic herbs | Hikal et al. [25] | |
| Maceration | Graphene Oxide-Based Extraction to adsorbs compounds | Oligosaccharides from fruit peels (inositol) | Ranjha et al. [26] | |
| Mechanical Methods | Physical pressure | Ultrasound-Assisted Extraction | Essential oils from citrus peels (limonene) | Chen et al. [27] |
| Grinding/Milling | Pulsed Electric Field Extraction | Antioxidants from fruit peels (quercetin) | Kaleem et al. [28] | |
| Shaking during maceration to boost extraction yield | Subcritical Water Extraction | Phenolic compounds from various food sources | Gbashi et al. [29], Beya et al. [30] and Kathiman et al. [31] |
|
| Heat-Based Methods | Heat Soxhlet Extraction by combining heat with solvents | Microwave-Assisted Extraction (Heating) | Carotenoids from vegetable waste (lycopene) | Anton et al. [32] and Vinatoru et al. [33] |
| Steam Distillation | Ohmic Heating Extraction | Volatile compounds Capsaicinoids from pepper waste (capsaicin) |
Hikal et al. [25] | |
| Chemical Methods | Acid/Base Hydrolysis | Enzyme-Assisted Extraction | Peptides (antioxidant peptides) | Costa et al. [34] and Prokopov et al. [35] |
| Aqueous Two-Phase for two immiscible liquids | Ionic Liquid Extraction | Alkaloids from agricultural waste (caffeine) | Costa et al. [34] | |
| Solid-Phase Methods | Solid-Phase Microextraction to absorbs compounds | Molecularly Imprinted Polymers (MIPs) for selective binding. | Polysaccharides from plant residues (β-glucans) | Gbashi et al. [29], Beya et al. [30] and Kathiman et al. [31] |
| Solid-Phase Extraction for compound adsorption | Graphene Oxide-Based Extraction for compound adsorption | Oligosaccharides from fruit peels (inositol) | Ranjha et al. [26] | |
| Other Methods | Steam Distillation and condensation | Supercritical Fluid Extraction (SFE) | Essential oils from various plant sources | Hikal et al. [25] and Kainat et al. [36] |
| Pressurized Liquid Extraction (PLE) or Accelerated Solvent Extraction (ASE) Combines solvent, high pressure, and temperature |
Enzyme-Assisted Extraction | Proteins and enzymes from agricultural waste | Costa et al. [34] | |
| Liquid–liquid extraction Separation of immiscible liquids | Instant Controlled Pressure Drop Technology for auto-vaporization and organic product development | Heat-sensitive food granule powder extraction | Kainat et al. [36] | |
| Solid-Phase Microextraction | Enzyme-Assisted Extraction | Active compounds from plant matter | Yang et al. [37] and Reshmitha et al. [38] | |
| Pressurized Liquid Extraction | Supercritical Fluids | Replacing organic solvents in various procedures | Kainat et al. [36] and Begić et al. [39] |
Several factors, including the prolonged extraction times, heavy reliance on hazardous solvents, and energy- and labor-intensive processes that affect solvent extraction methods’ repeatability and scalability, make conventional extraction techniques expensive, non-sustainable, and frequently unreliable. Conversely, advanced green extraction technologies are emerging that aim at maximizing the recovery of bioactive compounds while enhancing sustainability and efficiency thanks to rapid extraction rates, a reduced use of solvents, and low-energy demanding processes [40]. However, despite the reliance and efficiency of green and eco-friendly extraction methods, some of them still require further improvements and adjustments [41]. In fact, the inconsistency in the agricultural waste composition and physicochemical properties requires a substrate-driven tailoring of the enzymatic activities or microbial treatments and their associated process parameters, such as temperature, pH, substrate concentration, and reaction time.
4. Biological Methods
In the effort to extract valuable molecules from agricultural waste outputs, researchers have investigated both biological and non-biological methods. Conventional non-biological techniques depend on mechanical procedures like maceration and distillation, solvents, and thermal factors, while novel non-biological techniques use supercritical fluids, eutectic solvents, and ionic liquids. These methods’ shortcomings include low selectivity and specificity as well as the possibility of destroying thermo- or otherwise labile substances. Moreover, the complex cell walls in plant matrices may greatly hinder extraction efficiencies. To overcome this issue, various biomass pre-treatments have been developed, including physical, chemical, and physicochemical treatments, to overcome cell wall recalcitrance. Biological methods, applied as pre-treatments before extraction, are based on the exploitation of microbes or enzymes thereof to hydrolyze cell walls, thereby increasing permeability and compound recovery, which is, however, highly dependent on the characteristics of the agricultural waste material [42].
4.1. Microorganism-Driven Recovery of Bioactive Compounds
Numerous microorganisms, including both naturally occurring and genetically modified bacteria, yeasts, and filamentous fungi, have been used in fermentation processes that allowed the release of various bioactive chemicals from agro-industrial waste, including phenolic compounds, prebiotic oligomers, and β-glucan (Table 3). However, microbial fermentation can also be the source of valuable bioactive compounds, as in the case of carotenoids produced by several types of microorganisms in a process defined as bioconversion [43][44]. Microbial fermentation processes can be subdivided into two main types: solid-state fermentation (SSF) and submerged fermentation (SmF); the choice between the two mainly depends on the type of microorganism in use, on the target compound, and on the vegetable matrix in which it is contained [45]. In SmF, microorganisms are cultivated in a liquid medium in which the target compounds are released and subsequently collected using a separation process. The main drawback of SmF is represented by the dilution of the target products in the growth medium. However, SmF provides the operator with a perfect control over the fermentation conditions, and is ideally suited for microorganisms that have high moisture requirements [46]. In contrast, SSF exploits low free-water, solid substrates to grow microorganisms. The target compounds are released in the solid matrix and subsequently separated and extracted. The advantage of this approach includes the obtainment of more concentrated bioactive compounds, decreased wastewater generation, and high volumetric productivity. Both fermentation conditions require parameter optimization, which can be obtained through response surface methodology or one-factor-at-a-time approaches [10].
Table 3. Microbial fermentation-driven bioactive compound recovery from agro-industrial waste.
| Bioactive Compound | Substrate | Microorganism | Fermentation Process 1 | References |
|---|---|---|---|---|
| Phenolic Compounds | Black rice bran | Aspergillus awamori and Aspergillus oryzae | SSF | Shin et al. [47] |
| Apple peels | Aspergillus spp. (A. niger ZDM2 and A. tubingensis ZDM1) | SSF | Gulsunoglu et al. [43] | |
| Cauliflower | Bacillus subtilis | SmF | Doria et al. [48] | |
| Prebiotic Oligomers | Brewer’s spent grain | Trichoderma reesei B. subtilis (engineered strain) |
SmF | Amorim et al. [49][50] |
| Pectin | Citrus peel waste | Pichia pastoris (engineered strain) |
SmF | Yang et al. [51] |
| β-Glucan | Sugarcane straw | Lasiodiplodia Theobromae CCT 3966 | SmF | Abdeshahian et al. [52] |
| Soybean molasses | Lasiodiplodia theobromae MMPI | SmF | Acosta et al. [53] |
1 SmF: Submerged fermentation; SSF: Solid-state fermentation.
4.2. Microbial Enzymes
Microbial enzymes, including proteases, lipases, and several types of carbohydrases, play a pivotal role in recovering bioactive compounds. As already pointed out, when they are produced through the fermentation of agro-industrial wastes, they offer an economic advantage over synthetic or commercial enzymes, in particular when crude extracts obtained from such microbial factories are directly used as a source of enzymes; the inclusion of purification steps might, in fact, affect the economic viability of the process. For instance, Zanutto-Elgui et al. [54] used partially purified proteases produced by Aspergillus spp. through the SSF of wheat bran to derive bioactive peptides from bovine and goat milk; conversely, Doria et al. [48] explored the use of crude culture broth of wild-type and engineered Bacillus subtilis, overexpressing cellulases and xylanases, to recover phenolic compounds from cauliflower, identifying a positive correlation between the amount of Bacillus-originated cellulolytic enzymes in the culture broth and the bioactive compound yield. Thus, pre-treatment with culture broth is an effective and sustainable method for extracting bioactives from waste material.
4.3. Enzyme-Assisted Extraction (EAE)
EAE stands as an innovative approach to the recovery of various biomolecules. This technique involves the incorporation of cell wall-degrading or -weakening enzymes into the extraction medium, allowing the solvent to efficiently reach and extract the active constituents [34][55]. EAE methods offer an environmentally friendly means to facilitate the release of bioactive elements from agro-industrial waste materials, and wide adaptability as effective pre-treatment strategies, presenting several advantages over conventional pre-treatment techniques. Degradative enzymes can be seamlessly integrated with various downstream extraction techniques resulting in cleaner and more sustainable extraction processes. The combination of multiple approaches offers numerous advantages, such as improved product quality, reduced manufacturing costs, decreased solvent usage, and enhanced extraction yields [56]. Hydrolytic enzymes, whether in free or immobilized forms, can encompass a diverse range, including hemicellulases (endoxylanases and β-xylosidases), cellulases (endoglucanases, cellobiohydrolases, and β-glucosidases), and pectinases (polygalacturonases and pectinesterases), each playing an indispensable role in EAE. Proteases, amylases, lipases, and other hydrolyzing enzymes may also help to release valuable and scarce secondary metabolites from cellular constituents [7]. However, as often happens with biological tools, various operating variables, such as pH, temperature, substrate and enzyme concentrations, solid/liquid ratio, substrate particle size, and reaction duration, can influence the effectiveness of enzymatic activity [42].
A significant benefit of employing enzymes in industrial extraction is that they can be considered catalysts and be immobilized on solid supports to be recovered after use. Enzyme immobilization is a cost-effective strategy in large-scale applications; it allows enzymes to be employed in multiple cycles, while maintaining their catalytic activity and selectivity, reducing costs, and encouraging the recyclable use of the biological materials [57]. Magnetic nanoparticles as potential carriers for enzyme immobilization open up exciting prospects for enzyme-based extraction technologies. Moreover, the abundant supply of agro-industrial waste can be harnessed as microbial feedstock for enzyme production, thereby enhancing the cost-effectiveness of EAE.
5. Artificial Intelligence for the Digitization of Extraction Processes
5.1. Integration of AI/ML in the Recovery Process
Artificial intelligence (AI) can be integral to enhancing the efficiency and quality of the extraction process, making significant contributions across three key stages of application: pre-extraction, during extraction, and post extraction (Figure 3).
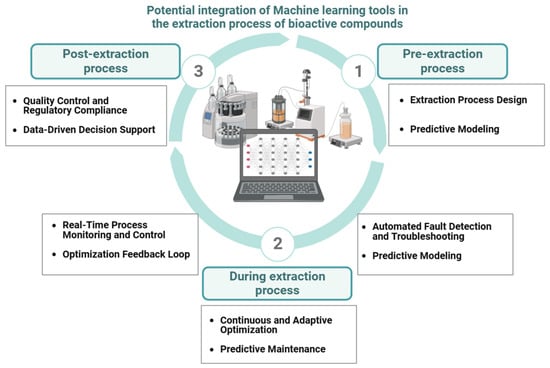
Figure 3. AI algorithm implementation for extraction (bio)process digitization (created with Biorender.com).
-
Pre-extraction stage: AI-based process design is a crucial component at this stage. Artificial intelligence models examine and screen historical data to determine the optimal solvent type, concentration, and temperature for a specific plant waste, microbe, enzyme, and target molecule. This analysis results in a customized extraction plan that maximizes yield and quality, establishing the conditions for a more efficient process [10]. Moreover, predictive modeling can anticipate the yield and titer of the extracted compounds by combining historical data and real-time sensor data [10][58].
-
During the extraction process: At this phase, AI models can handle enormous amounts of data and assist in real-time monitoring and process control. Sensors continuously monitor critical parameters like pressure, temperature, and solvent flow rate [59][60], while AI algorithms examine real-time data and autonomously adjust conditions to maintain efficiency and consistency [60], correcting deviations from expected patterns [58][61]. A related area of potential application for AI models is continuous and adaptive optimization. Here, the algorithm learns from new data, identifying patterns and suggesting process refinements and efficiency enhancements in the extraction process [62]. Finally, machine learning methods can be used in large-scale extraction/production plants in processes of predictive maintenance of equipment, limiting downtime and ensuring continuous output [59][63].
-
Post-extraction stage: AI can help to interpret extraction data and assist in decision-making for additional processing, purification, and quality control processes at the final phase of bioactive molecule recovery, providing data-driven decision support and processing [58]. Moreover, AI models may assist in quality control and regulatory compliance. They ensure that the final compound satisfies safety, quality, or regulatory requirements and reduce the need for manual examinations [64].
5.2. Standard Workflow of Machine Learning
Among the various AI approaches, machine learning (ML) methods have been increasingly used in food processing and bioactive compound extraction [65][66]. They can treat non-linearly coupled complex data, perform modelling, and generate the classification, prediction, or optimization of different responses [10].
ML algorithms tune the model’s internal parameters so that it fits a training dataset (learning phase), and when the training is over, the model can be used to generate accurate predictions on fresh input data (prediction phase) [67][68]. While there is a wealth of ML models, each with its rather complicated details and preferred application domains, their integration entails a general and standardized workflow which deserves a brief description (Figure 4).
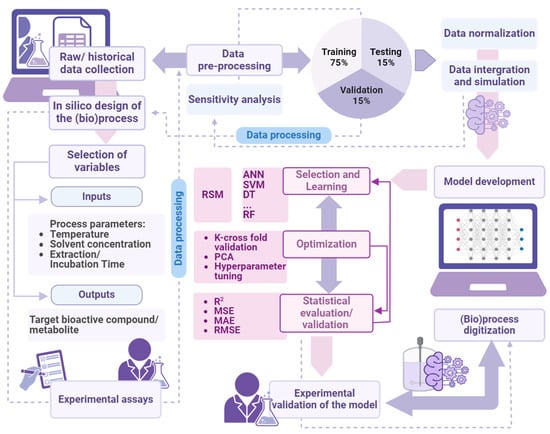
Figure 4. Digitized extraction (bio)process development and manufacturing using machine learning (created with Biorender.com).
The main workflow tasks are as follows:
- (a)
-
Data collection
It involves extracting relevant and representative data to train, validate, and test the model. In case of bioactive extraction processes, these data must be obtained through extensive experimental measurements.
- (b)
-
Data preprocessing
It consists in cleaning and preprocessing the data, possibly handling missing values, dealing with outliers, scaling values, encoding categorical variables, and, above all, reducing the dataset while retaining its essential information (dimension reduction).
- (c)
-
Data splitting
This step consists in partitioning the data into training (70%), validation (15%), and testing (15%) sets, the first used in the learning phase, the second to tune and confirm the model, and the third to verify its performance.
- (d)
-
Model selection
The selection of the right model, which depends on the specific task it is designed to solve, is a key point in the process. Model selection is often a matter of trial and error, and sometimes hybrid approaches (mixing statistical and AI models) must be adopted. For instance, an ANN model was used to optimize the extraction of catechins from green tea leaves [9].
- (e)
-
Model development and optimization
This is the step in which the model is trained, usually in several iterative runs. The effectiveness of the training and the quality of the ML model depend, apart from the training dataset, on the right values of its control parameters (called hyperparameters in this domain), which are set before the training phase and whose fine-tuning is pivotal for optimal model performance [69]. The correct choice of hyperparameters helps to avoid overfitting, a common disease of ML, yielding models so perfectly fit to the training set that do not generalize to other inputs.
- (f)
-
Model Evaluation
Evaluation involves model validation and test, which are similar but distinct activities. The first, sometimes considered optional, aims at selecting a model (and its hyperparameters) among several possible choices, assessing its performances on a validation dataset; the second, mandatory, verifies the quality of the model against a test dataset.
In conclusion, the integration of AI into biomass-refining plants is essential for enhancing economic return and operating conditions, boosting the attractiveness of AI in industrial settings. Automation and AI tools can, in fact, accelerate large-scale bioactive compound production and extraction.
However, currently, the integration of AI to the recovery process is still challenging. Even when AI promises to significantly increase control, quality, and efficiency throughout the extraction process, a smooth and successful integration into the plant waste recovery process depends on finding innovative solutions to overcome these limitations. Collaboration with domain specialists is essential to successfully manage the intrinsic complexities.
References
- Chauhan, C.; Dhir, A.; Akram, M.U.; Salo, J. Food Loss and Waste in Food Supply Chains. A Systematic Literature Review and Framework Development Approach. J. Clean. Prod. 2021, 295, 126438.
- Sadh, P.K.; Duhan, S.; Duhan, J.S. Agro-Industrial Wastes and Their Utilization Using Solid State Fermentation: A Review. Bioresour. Bioprocess. 2018, 5, 1.
- Lopes, F.C.; Ligabue-Braun, R. Agro-Industrial Residues: Eco-Friendly and Inexpensive Substrates for Microbial Pigments Production. Front. Sustain. Food Syst. 2021, 5, 589414.
- Ferri, M.; Rodríguez, Ó.; Tassoni, A. Editorial: New Green Extraction Methods for the Sustainable Recovery of Functional Plant Secondary Metabolites. Front. Plant Sci. 2023, 14, 1186180.
- Heemann, A.C.W.; Heemann, R.; Kalegari, P.; Spier, M.R.; Santin, E. Enzyme-Assisted Extraction of Polyphenols from Green Yerba Mate. Braz. J. Food Technol. 2019, 22, e2017222.
- Wang, T.; Lü, X. Overcome Saccharification Barrier. In Advances in 2nd Generation of Bioethanol Production; Elsevier: Amsterdam, The Netherlands, 2021; pp. 137–159. ISBN 9780128188620.
- Marathe, S.J.; Jadhav, S.B.; Bankar, S.B.; Kumari Dubey, K.; Singhal, R.S. Improvements in the Extraction of Bioactive Compounds by Enzymes. Curr. Opin. Food Sci. 2019, 25, 62–72.
- Lemes, A.C.; Egea, M.B.; de Oliveira Filho, J.G.; Gautério, G.V.; Ribeiro, B.D.; Coelho, M.A.Z. Biological Approaches for Extraction of Bioactive Compounds from Agro-Industrial By-Products: A Review. Front. Bioeng. Biotechnol. 2022, 9, 802543.
- Shishodia, A.; Kumar, K.; Manna, M.S. Modeling for the Efficient Separation of Bio-Active Catechins from Green Tea Leaves. Sep. Sci. Technol. 2017, 52, 671–678.
- Alloun, W.; Berkani, M.; Benaissa, A.; Shavandi, A.; Gares, M.; Danesh, C.; Lakhdari, D.; Ghfar, A.A.; Chaouche, N.K. Waste Valorization as Low-Cost Media Engineering for Auxin Production from the Newly Isolated Streptomyces Rubrogriseus AW22: Model Development. Chemosphere 2023, 326, 138394.
- Velliquette, R.A.; Fast, D.J.; Maly, E.R.; Alashi, A.M.; Aluko, R.E. Enzymatically Derived Sunflower Protein Hydrolysate and Peptides Inhibit NFκB and Promote Monocyte Differentiation to a Dendritic Cell Phenotype. Food Chem. 2020, 319, 126563.
- Hemker, A.K.; Nguyen, L.T.; Karwe, M.; Salvi, D. Effects of Pressure-Assisted Enzymatic Hydrolysis on Functional and Bioactive Properties of Tilapia (Oreochromis niloticus) by-Product Protein Hydrolysates. LWT 2020, 122, 109003.
- Huang, M.; Wang, H.; Xu, X.; Lu, X.; Song, X.; Zhou, G. Effects of Nanoemulsion-Based Edible Coatings with Composite Mixture of Rosemary Extract and ε-Poly-L-Lysine on the Shelf Life of Ready-to-Eat Carbonado Chicken. Food Hydrocoll. 2020, 102, 105576.
- Lovegrove, A.; Edwards, C.H.; De Noni, I.; Patel, H.; El, S.N.; Grassby, T.; Zielke, C.; Ulmius, M.; Nilsson, L.; Butterworth, P.J.; et al. Role of Polysaccharides in Food, Digestion, and Health. Crit. Rev. Food Sci. Nutr. 2017, 57, 237–253.
- Fortunati, E.; Luzi, F.; Puglia, D.; Torre, L. Extraction of Lignocellulosic Materials from Waste Products. In Multifunctional Polymeric Nanocomposites Based on Cellulosic Reinforcements; William Andrew Publishing: Norwich, NY, USA, 2016; pp. 1–38. ISBN 9780323442480.
- Tosh, S.M.; Brummer, Y.; Miller, S.S.; Regand, A.; Defelice, C.; Duss, R.; Wolever, T.M.S.; Wood, P.J. Processing Affects the Physicochemical Properties of β-Glucan in Oat Bran Cereal. J. Agric. Food Chem. 2010, 58, 7723–7730.
- Anarjan, N.; Jouyban, A. Preparation of Lycopene Nanodispersions from Tomato Processing Waste: Effects of Organic Phase Composition. Food Bioprod. Process. 2017, 103, 104–113.
- Caseiro, M.; Ascenso, A.; Costa, A.; Creagh-Flynn, J.; Johnson, M.; Simões, S. Lycopene in Human Health. LWT 2020, 127, 109323.
- Dave, D.; Routray, W. Current Scenario of Canadian Fishery and Corresponding Underutilized Species and Fishery Byproducts: A Potential Source of Omega-3 Fatty Acids. J. Clean. Prod. 2018, 180, 617–641.
- Guimarães, R.M.; Pimentel, T.C.; de Rezende, T.A.M.; de Silva, J.S.; Falcão, H.G.; Ida, E.I.; Egea, M.B. Gluten-Free Bread: Effect of Soy and Corn Co-Products on the Quality Parameters. Eur. Food Res. Technol. 2019, 245, 1365–1376.
- Nogueira, G.F.; de Oliveira, R.A.; Velasco, J.I.; Fakhouri, F.M. Methods of Incorporating Plant-Derived Bioactive Compounds into Films Made with Agro-Based Polymers for Application as Food Packaging: A Brief Review. Polymers 2020, 12, 2518.
- de Oliveira Filho, J.G.; Braga, A.R.C.; de Oliveira, B.R.; Gomes, F.P.; Moreira, V.L.; Pereira, V.A.C.; Egea, M.B. The Potential of Anthocyanins in Smart, Active, and Bioactive Eco-Friendly Polymer-Based Films: A Review. Food Res. Int. 2021, 142, 110202.
- Alongi, M.; Anese, M. Re-Thinking Functional Food Development through a Holistic Approach. J. Funct. Foods 2021, 81, 104466.
- Carrasco-Sandoval, J.; Rebolledo, P.; Peterssen-Fonseca, D.; Fischer, S.; Wilckens, R.; Aranda, M.; Henríquez-Aedo, K. A Fast and Selective Method to Determine Phenolic Compounds in Quinoa (Chenopodium Quinoa Will) Seeds Applying Ultrasound-Assisted Extraction and High-Performance Liquid Chromatography. Chem. Pap. 2021, 75, 431–438.
- Hikal, W.M.; Said-Al Ahl, H.A.H.; Tkachenko, K.G.; Bratovcic, A.; Szczepanek, M.; Rodriguez, R.M. Sustainable and Environmentally Friendly Essential Oils Extracted from Pineapple Waste. Biointerface Res. Appl. Chem. 2022, 12, 6833–6844.
- Ranjha, M.M.A.N.; Amjad, S.; Ashraf, S.; Khawar, L.; Safdar, M.N.; Jabbar, S.; Nadeem, M.; Mahmood, S.; Murtaza, M.A. Extraction of Polyphenols from Apple and Pomegranate Peels Employing Different Extraction Techniques for the Development of Functional Date Bars. Int. J. Fruit Sci. 2020, 20, S1201–S1221.
- Chen, X.; Li, X.; Zhu, X.; Wang, G.; Zhuang, K.; Wang, Y.; Ding, W. Optimization of Extrusion and Ultrasound-Assisted Extraction of Phenolic Compounds from Jizi439 Black Wheat Bran. Processes 2020, 8, 1153.
- Kaleem, M.; Ahmad, A.; Amir, R.M.; Raja, G.K. Ultrasound-Assisted Phytochemical Extraction Condition Optimization Using Response Surface Methodology from Perlette Grapes (Vitis vinifera). Processes 2019, 7, 749.
- Gbashi, S.; Adebo, O.A.; Piater, L.; Madala, N.E.; Njobeh, P.B. Subcritical Water Extraction of Biological Materials. Sep. Purif. Rev. 2017, 46, 21–34.
- Beya, M.M.; Netzel, M.E.; Sultanbawa, Y.; Smyth, H.; Hoffman, L.C. Plant-Based Phenolic Molecules as Natural Preservatives in Comminuted Meats: A Review. Antioxidants 2021, 10, 263.
- Kathiman, M.N.; Mudalip, S.K.A.; Gimbun, J. Effect of Encapsulation Agents on Antioxidant Activity and Moisture Content of Spray Dried Powder from Mahkota Dewa Fruit Extract. In IOP Conference Series: Materials Science and Engineering; IOP Publishing: Bristol, UK, 2020; Volume 991, p. 012040. Available online: https://iopscience.iop.org/article/10.1088/1757-899X/991/1/012040 (accessed on 31 December 2023).
- Anton, I.; Húth, B.; Füller, I.; Rózsa, L.; Holló, G.; Zsolnai, A. Effect of Single Nucleotide Polymorphisms on Intramuscular Fat Content in Hungarian Simmental Cattle. Asian-Australasian J. Anim. Sci. 2018, 31, 1415–1419.
- Vinatoru, M.; Mason, T.J.; Calinescu, I. Ultrasonically Assisted Extraction (UAE) and Microwave Assisted Extraction (MAE) of Functional Compounds from Plant Materials. TrAC Trends Anal. Chem. 2017, 97, 159–178.
- Costa, J.R.; Tonon, R.V.; Cabral, L.; Gottschalk, L.; Pastrana, L.; Pintado, M.E. Valorization of Agricultural Lignocellulosic Plant Byproducts through Enzymatic and Enzyme-Assisted Extraction of High-Value-Added Compounds: A Review. ACS Sustain. Chem. Eng. 2020, 8, 13112–13125.
- Prokopov, T.; Nikolova, M.; Dobrev, G.; Taneva, D. Enzyme-Assisted Extraction of Carotenoids from Bulgarian Tomato Peels. Acta Aliment. 2017, 46, 84–91.
- Kainat, S.; Arshad, M.S.; Khalid, W.; Zubair Khalid, M.; Koraqi, H.; Afzal, M.F.; Noreen, S.; Aziz, Z.; Al-Farga, A. Sustainable Novel Extraction of Bioactive Compounds from Fruits and Vegetables Waste for Functional Foods: A Review. Int. J. Food Prop. 2022, 25, 2457–2476.
- Yang, X.; Zhang, Y.; Pang, H.; Yuan, S.; Wang, X.; Hu, Z.; Zhou, Q.; He, Y.; Yan, Y.; Xu, L. Codisplay of Rhizopus Oryzae and Candida Rugosa Lipases for Biodiesel Production. Catalysts 2021, 11, 421.
- Reshmitha, T.R.; Thomas, S.; Geethanjali, S.; Arun, K.B.; Nisha, P. DNA and Mitochondrial Protective Effect of Lycopene Rich Tomato (Solanum lycopersicum L.) Peel Extract Prepared by Enzyme Assisted Extraction against H2O2 Induced Oxidative Damage in L6 Myoblasts. J. Funct. Foods 2017, 28, 147–156.
- Begić, S.; Horozić, E.; Alibašić, H.; Bjelić, E.; Seferović, S.; Kozarević, E.C.; Ibišević, M.; Zukić, A.; Karić, E.; Softić, M. Antioxidant Capacity and Total Phenolic and Flavonoid Contents of Methanolic Extracts of Urtica Dioica L. by Different Extraction Techniques. Int. Res. J. Pure Appl. Chem. 2020, 21, 207–214.
- Mena-García, A.; Ruiz-Matute, A.I.; Soria, A.C.; Sanz, M.L. Green Techniques for Extraction of Bioactive Carbohydrates. TrAC Trends Anal. Chem. 2019, 119, 115612.
- Pocha, C.K.R.; Chia, W.Y.; Chew, K.W.; Munawaroh, H.S.H.; Show, P.L. Current Advances in Recovery and Biorefinery of Fucoxanthin from Phaeodactylum tricornutum. Algal Res. 2022, 65, 102735.
- Hernández Becerra, E.; De Jesús Pérez López, E.; Zartha Sossa, J.W. Recovery of Biomolecules from Agroindustry by Solid-Liquid Enzyme-Assisted Extraction: A Review. Food Anal. Methods 2021, 14, 1744–1777.
- Gulsunoglu, Z.; Purves, R.; Karbancioglu-Guler, F.; Kilic-Akyilmaz, M. Enhancement of Phenolic Antioxidants in Industrial Apple Waste by Fermentation with Aspergillus Spp. Biocatal. Agric. Biotechnol. 2020, 25, 101562.
- Sinha, S.; Singh, G.; Arora, A.; Paul, D. Carotenoid Production by Red Yeast Isolates Grown in Agricultural and “Mandi” Waste. Waste Biomass Valorization 2021, 12, 3939–3949.
- Bhanja Dey, T.; Chakraborty, S.; Jain, K.K.; Sharma, A.; Kuhad, R.C. Antioxidant Phenolics and Their Microbial Production by Submerged and Solid State Fermentation Process: A Review. Trends Food Sci. Technol. 2016, 53, 60–74.
- Bagewadi, Z.K.; Mulla, S.I.; Ninnekar, H.Z. Optimization of Endoglucanase Production from Trichoderma Harzianum Strain HZN11 by Central Composite Design under Response Surface Methodology. Biomass Convers. Biorefinery 2018, 8, 305–316.
- Shin, H.Y.; Kim, S.M.; Lee, J.H.; Lim, S.T. Solid-State Fermentation of Black Rice Bran with Aspergillus awamori and Aspergillus oryzae: Effects on Phenolic Acid Composition and Antioxidant Activity of Bran Extracts. Food Chem. 2019, 272, 235–241.
- Doria, E.; Buonocore, D.; Marra, A.; Bontà, V.; Gazzola, A.; Dossena, M.; Verri, M.; Calvio, C. Bacterial-Assisted Extraction of Bioactive Compounds from Cauliflower. Plants 2022, 11, 816.
- Amorim, C.; Silvério, S.C.; Rodrigues, L.R. One-Step Process for Producing Prebiotic Arabino-Xylooligosaccharides from Brewer’s Spent Grain Employing Trichoderma Species. Food Chem. 2019, 270, 86–94.
- Amorim, C.; Silvério, S.C.; Silva, S.P.; Coelho, E.; Coimbra, M.A.; Prather, K.L.J.; Rodrigues, L.R. Single-Step Production of Arabino-Xylooligosaccharides by Recombinant Bacillus Subtilis 3610 Cultivated in Brewers’ Spent Grain. Carbohydr. Polym. 2018, 199, 546–554.
- Yang, G.; Tan, H.; Li, S.; Zhang, M.; Che, J.; Li, K.; Chen, W.; Yin, H. Application of Engineered Yeast Strain Fermentation for Oligogalacturonides Production from Pectin-Rich Waste Biomass. Bioresour. Technol. 2020, 300, 122645.
- Abdeshahian, P.; Ascencio, J.J.; Philippini, R.R.; Antunes, F.A.F.; dos Santos, J.C.; da Silva, S.S. Utilization of Sugarcane Straw for Production of β-Glucan Biopolymer by Lasiodiplodia Theobromae CCT 3966 in Batch Fermentation Process. Bioresour. Technol. 2020, 314, 123716.
- Acosta, S.B.P.; Marchioro, M.L.K.; Santos, V.A.Q.; Calegari, G.C.; Lafay, C.B.B.; Barbosa-Dekker, A.M.; Dekker, R.F.H.; da Cunha, M.A.A. Valorization of Soybean Molasses as Fermentation Substrate for the Production of Microbial Exocellular β-Glucan. J. Polym. Environ. 2020, 28, 2149–2160.
- Zanutto-Elgui, M.R.; Vieira, J.C.S.; do Prado, D.Z.; Buzalaf, M.A.R.; de Padilha, P.M.; Elgui de Oliveira, D.; Fleuri, L.F. Production of Milk Peptides with Antimicrobial and Antioxidant Properties through Fungal Proteases. Food Chem. 2019, 278, 823–831.
- Nadar, S.S.; Rao, P.; Rathod, V.K. Enzyme Assisted Extraction of Biomolecules as an Approach to Novel Extraction Technology: A Review. Food Res. Int. 2018, 108, 309–330.
- Picot-Allain, C.; Mahomoodally, M.F.; Ak, G.; Zengin, G. Conventional versus Green Extraction Techniques—A Comparative Perspective. Curr. Opin. Food Sci. 2021, 40, 144–156.
- Basso, A.; Serban, S. Industrial Applications of Immobilized Enzymes—A Review. Mol. Catal. 2019, 479, 110607.
- Khanal, S.K.; Tarafdar, A.; You, S. Artificial Intelligence and Machine Learning for Smart Bioprocesses. Bioresour. Technol. 2023, 375, 128826.
- Caudai, C.; Galizia, A.; Geraci, F.; Le Pera, L.; Morea, V.; Salerno, E.; Via, A.; Colombo, T. AI Applications in Functional Genomics. Comput. Struct. Biotechnol. J. 2021, 19, 5762–5790.
- Rai, R.; Tiwari, M.K.; Ivanov, D.; Dolgui, A. Machine Learning in Manufacturing and Industry 4.0 Applications. Int. J. Prod. Res. 2021, 59, 4773–4778.
- Sarker, K.U.; Saqib, M.; Hasan, R.; Mahmood, S.; Hussain, S.; Abbas, A.; Deraman, A. A Ranking Learning Model by K-Means Clustering Technique for Web Scraped Movie Data. Computers 2022, 11, 158.
- Mowbray, M.; Savage, T.; Wu, C.; Song, Z.; Cho, B.A.; Del Rio-Chanona, E.A.; Zhang, D. Machine Learning for Biochemical Engineering: A Review. Biochem. Eng. J. 2021, 172, 108054.
- Kavasidis, I.; Lallas, E.; Gerogiannis, V.C.; Charitou, T.; Karageorgos, A. Predictive Maintenance in Pharmaceutical Manufacturing Lines Using Deep Transformers. In Procedia Computer Science; Elsevier: Amsterdam, The Netherlands, 2023; Volume 220, pp. 576–583.
- Wainaina, S.; Taherzadeh, M.J. Automation and Artificial Intelligence in Filamentous Fungi-Based Bioprocesses: A Review. Bioresour. Technol. 2023, 369, 128421.
- Al-Sammarraie, M.A.J.; Gierz, Ł.; Przybył, K.; Koszela, K.; Szychta, M.; Brzykcy, J.; Baranowska, H.M. Predicting Fruit’s Sweetness Using Artificial Intelligence—Case Study: Orange. Appl. Sci. 2022, 12, 8233.
- Gomes, V.; Reis, M.S.; Rovira-Más, F.; Mendes-Ferreira, A.; Melo-Pinto, P. Prediction of Sugar Content in Port Wine Vintage Grapes Using Machine Learning and Hyperspectral Imaging. Processes 2021, 9, 1241.
- Taşpınar, H.; Elik, A.; Kaya, S.; Altunay, N. Optimization of Green and Rapid Analytical Procedure for the Extraction of Patulin in Fruit Juice and Dried Fruit Samples by Air-Assisted Natural Deep Eutectic Solvent-Based Solidified Homogeneous Liquid Phase Microextraction Using Experimental Design And Computational Chemistry Approach. Food Chem. 2021, 358, 129817.
- Kumbhar, A.; Dhawale, P.G.; Kumbhar, S.; Patil, U.; Magdum, P. A Comprehensive Review: Machine Learning and Its Application in Integrated Power System. Energy Rep. 2021, 7, 5467–5474.
- Hertel, L.; Collado, J.; Sadowski, P.; Ott, J.; Baldi, P. Sherpa: Robust Hyperparameter Optimization for Machine Learning. SoftwareX 2020, 12, 100591.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
597
Revisions:
2 times
(View History)
Update Date:
07 Mar 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




