Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Gratiela GRADISTEANU Pircalabioru | -- | 3031 | 2024-03-05 11:34:38 | | | |
| 2 | Peter Tang | Meta information modification | 3031 | 2024-03-06 06:44:30 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Gradisteanu Pircalabioru, G.; Musat, M.; Elian, V.; Iliescu, C. Liquid Biopsy in Type 2 Diabetes. Encyclopedia. Available online: https://encyclopedia.pub/entry/55870 (accessed on 07 February 2026).
Gradisteanu Pircalabioru G, Musat M, Elian V, Iliescu C. Liquid Biopsy in Type 2 Diabetes. Encyclopedia. Available at: https://encyclopedia.pub/entry/55870. Accessed February 07, 2026.
Gradisteanu Pircalabioru, Gratiela, Madalina Musat, Viviana Elian, Ciprian Iliescu. "Liquid Biopsy in Type 2 Diabetes" Encyclopedia, https://encyclopedia.pub/entry/55870 (accessed February 07, 2026).
Gradisteanu Pircalabioru, G., Musat, M., Elian, V., & Iliescu, C. (2024, March 05). Liquid Biopsy in Type 2 Diabetes. In Encyclopedia. https://encyclopedia.pub/entry/55870
Gradisteanu Pircalabioru, Gratiela, et al. "Liquid Biopsy in Type 2 Diabetes." Encyclopedia. Web. 05 March, 2024.
Copy Citation
As the burden of type 2 diabetes (T2D) continues to escalate globally, there is a growing need for novel, less-invasive biomarkers capable of early diabetes detection and monitoring of disease progression. Liquid biopsy, recognized for its minimally invasive nature, is increasingly being applied beyond oncology, and nevertheless shows its potential when the collection of the tissue biopsy is not possible. This diagnostic approach involves utilizing liquid biopsy markers such as cell-free nucleic acids, extracellular vesicles, and diverse metabolites for the molecular diagnosis of T2D and its related complications.
type 2 diabetes
liquid biopsy
exosomes
miRNA
molecular diagnostic
1. Introduction
The rapid increase in prevalence of type 2 diabetes mellitus (T2D) qualifies it as one of the fastest-growing global health emergencies of the 21st century [1]. T2D is a chronic condition necessitating continuous management and attention to avert its extensive health consequences, including myocardial infarction, cardiac failure, blindness, kidney failure, stroke, and amputation.
T2D is a multifactorial disease resulting from a combination of factors, including inadequate beta-pancreatic cell (β-cell) function, reduced insulin sensitivity, and chronic inflammation that precede the diabetes onset with up to 15 years. Currently, the diagnosis of T2D is made typically once hyperglycemia has become evident, a limitation that impedes early disease detection and precludes the possibility of timely interventions aimed at preserving β-cell mass and improving patient outcomes. Liquid biopsy can be a promising solution to this challenge, being useful in the diagnostic, management, and prognostic of the disease and providing a deeper understanding of the disease’s pathophysiology.
Early detection and intervention are crucial for preventing the progression of T2D and reducing the risk of complications such as nephropathy, cardiovascular disease, neuropathy, or retinopathy. Even though biomarkers such as glycemia or HbA1c are widely used for diagnosing T2D, presently, there is no biomarker that is able to predict the ongoing destruction of beta cells in the pancreas in the early stages of the disease as opposed to T1D, where specific autoantibodies against pancreatic islet cells or insulin are often present before the onset of symptoms and individuals at increased risk or in the early stages of T1D can be timely identified.
On the other hand, liquid biopsy shows its potential in the prognosis, diagnosis, and monitoring of cancer [2][3][4]. The use of a small amount of “body fluids” (usually blood), containing biomarkers that can provide relevant information for diagnostic and monitoring, present some relevant advantages such as reduced invasiveness, easy protocol for sample collection, lower costs, and real-time information [5][6][7].
2. Liquid Biopsy Markers in T2D
Biomarkers that allow for early diagnosis and effective monitoring have the potential to lead to better patient-centric therapeutic decisions. In T2D, identifying the precise timing of critical events such as β-cell stress, de-differentiation, or death is essential for developing effective treatments [8]. However, the inaccessibility of pancreatic tissue in humans for longitudinal, minimally invasive monitoring represents a significant obstacle. To address this shortcoming, liquid biopsy could be employed for the detection of β-cell markers. Liquid biopsies encompass a diverse range of clinically informative elements. Blood samples can undergo analysis for metabolites, exosomes, and cell-free nucleic acids, each offering valuable numerical and molecular insights. Employment of liquid biopsy in T2D patients can provide valuable insight regarding T2D complications, cardiovascular disease (CVD), and cancer risk. An overview of the key elements and the impact of the liquid biopsy markers in T2D is presented in Figure 1.
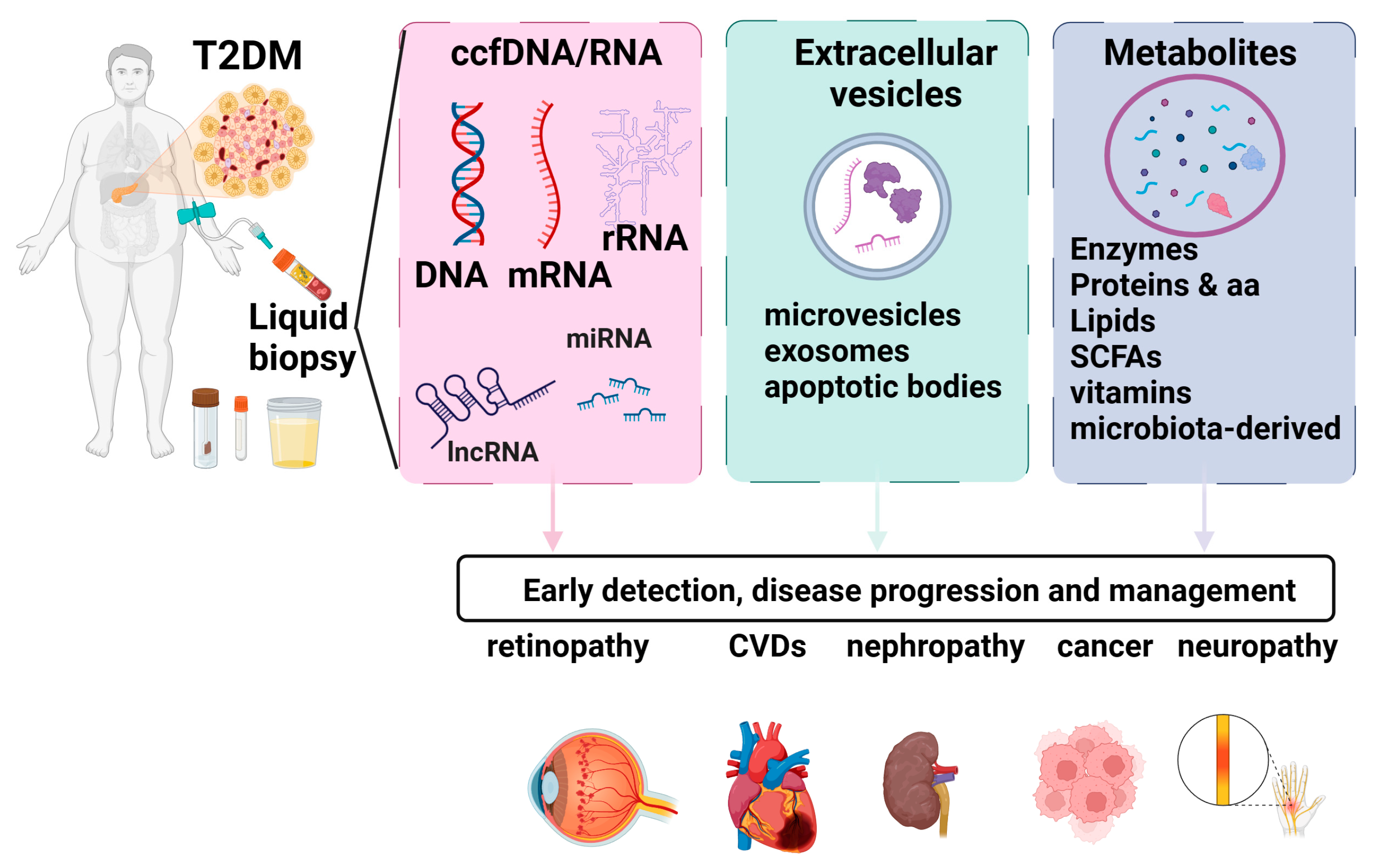
Figure 1. Liquid biopsy markers in T2D. Liquid biopsies encompass a diverse range of clinically informative elements. Blood samples can undergo analysis for metabolites, exosomes, and cell-free nucleic acids, each offering valuable numerical and molecular insights. Employment of liquid biopsy in T2D patients can provide valuable insight regarding T2D complications, cardiovascular disease (CVD), and cancer risk. Figure created using biorender.com.
2.1. Cell-Free Nucleic Acids
2.1.1. Circulating Cell-Free DNA (ccfDNA)
Circulating cell-free DNA (ccfDNA), also known as cell-free DNA (cfDNA), refers to short (≈160 nt) double-stranded DNA fragments found in various biological fluids, including blood, urine, saliva, and cerebrospinal fluid. Generally, ccfDNA is actively released by living cells or can result after cell death (apoptosis, necrosis) [9].
Several methods have been utilized to assess cfDNA, including droplet digital polymerase chain reaction (ddPCR), beads, emulsion, amplification, and magnetics (BEAMing), tagged-amplicon deep sequencing (TAm-Seq), whole-genome bisulfite sequencing (WGBS-Seq), whole-exome sequencing (WES), and whole-genome sequencing (WGS).
BEAMing proves to be a cost-effective and highly sensitive screening method for identifying established mutations. The method of integrating PCR with flow cytometry has the capability to identify alterations at levels as minimal as 0.01%, demonstrating excellent concordance with tissue testing [10].
Droplet digital polymerase chain reaction (ddPCR) is capable of detecting genomic material at levels as low as 0.01–1.0%, making it a valuable tool for identifying potentially rare mutations and quantifying copy number variants [11]. Nevertheless, its applicability is limited to assessing the presence of well-characterized sequences.
In patients with cancer, whole-exome sequencing offers a comprehensive analysis of all existing tumor mutations, allowing for the identification of potential oncogenes and tumor suppressor genes. However, its sensitivity may be comparatively lower than other methods due to the inclusion of exomic alterations. Despite this, it stands out for its cost-effectiveness and high yield [12].
Tagged-amplicon deep sequencing (TAm-Seq) enables a highly specific and sensitive analysis, achieving an accuracy of approximately 97%. It has the capability to detect DNA levels as low as 2% through the utilization of primers to tag and identify genomic sequences. TAm-Seq boasts a high sequencing throughput, leading to reduced sequencing time and cost, allowing for the simultaneous sequencing of millions of DNA molecules. However, it is essential for the desired sequence to be pre-characterized for the methodology to be effective [13].
Whole-genome sequencing (WGS) examines the entirety of the sample genome to identify well-characterized and detrimental alterations, along with variants of unknown significance. It holds significant potential for a thorough assessment of genetic mutations and, in case of T2D, could be used to identify cancer risk; however, its application is constrained by challenges in quality assurance, ethical considerations, time, and cost. The interpretation of results can be challenging outside of specialized centers [12].
Whole-genome bisulfite sequencing (WGBS-Seq) stands as the benchmark in DNA methylation analysis. It provides a measurement for each cytosine with exceptional accuracy. While it has the capability to uncover partially methylated domains in cancer cells, the sensitivity of this method can be compromised due to potential variations in DNA degradation [12].
Cell-free DNA (cfDNA)-based biomarkers have shown promise as minimally invasive options for the early diagnosis and monitoring of diabetes in various studies [14][15].
CcfDNA fragments retain characteristics indicative of their tissue of origin. Determining the ccfDNA tissue of origin can be performed using different approaches: the detection of distinct SNPs and/or genetic mutations, genome-wide methylation or 5hmC profiling, and fragmentation profiling by nucleosome footprints [16].
Adipose tissue dysfunction, characterized by hypertrophy, inflammation, and cell death, is closely associated with the development of insulin resistance—a key feature of T2D. Hence, the detection of cfDNA fragments associated with adipose tissue degeneration can provide insights into the mechanisms underlying insulin resistance [17].
Combining cfDNA concentration with tissue-specific epigenetic signatures may offer a comprehensive approach to assessing metabolic risk and disease progression. For instance, Lehmann-Werman et al. used methylation profiling to identify circulating DNA linked to different pathological processes: pancreatic β-cell DNA from patients with type 1 diabetes and exocrine pancreatic DNA from patients with pancreatic cancer [18].
Karaglani et al. evaluated the methylation profile of a panel of specific genes related to β-cells, including INS (insulin), GCK (glucokinase), IAPP (islet amyloid polypeptide-amylin), KCNJ11 (potassium inwardly rectifying channel subfamily J member 11), and ABCC8 (ATP binding cassette subfamily C member 8) and, based on the data generated, they built classifying predictive models using automated machine learning. INS, IAPP, GCK, and KCNJ11 levels differed significantly between T2D patients and healthy controls. Based on ccfDNA parameters, methylation data and demographical information, automated machine learning analysis generated biosignatures, including GCK, IAPP, and KCNJ11 methylation, with a very high discriminating performance of T2D from healthy individuals [19].
While human leukocyte antigen (HLA) typing is primarily used for assessing the risk of developing type 1 diabetes, there is ongoing research exploring potential associations between specific HLA alleles and the risk of diabetes-related complications. These complications may include diabetic nephropathy, retinopathy, and neuropathy. HLA typing, combined with other genetic and clinical information, may contribute to individualized risk assessments for diabetes complications. Identifying individuals at higher risk may allow for targeted monitoring and early interventions to prevent or manage complications. Ongoing research is exploring the complex interplay between genetics, including HLA alleles, and the development of diabetes and its complications. Advances in genomic medicine and personalized medicine may lead to more refined risk assessments and tailored therapeutic approaches. The influence of variations in the HLA genotype on the development of type 2 diabetes complications is not clearly understood. Recently, Birinci et al. (2022) identified the HLA variant HLA-DR7 to be associated with diabetic rethynopathy development [20]. The presence of the HLA-DQB1*0501 allele demonstrated a protective association against diabetic nephropathy (DN) in individuals of Han ethnicity in China [21].
Cell-free mitochondrial DNA (ccf-mtDNA) is produced and discharged from cells into the systemic circulation in response to cellular damage or stress. This release may occur passively through various forms of cell death or actively from living cells through processes that are not fully comprehended at present [22]. Increased blood ccf-mtDNA levels have been associated with T2D associated with coronary heart disease or cognitive impairment [23][24].
2.1.2. Circulating Cell-Free RNA (ccfRNA)
Human blood is home to a wide range of cfRNA species, including microRNA (miRNA), messenger RNA (mRNA), long-noncoding RNA(lncRNA), Piwi-interactingRNA (piRNA), circular RNA (circRNA), transfer RNA (tRNA), and miscellaneous other noncoding RNA molecules [25].
Advances in high-throughput sequencing and bioinformatics have improved the ability to profile and analyze cfRNA. Emerging single-cell RNA sequencing (scRNA-seq) technologies enabled the study of cfRNA heterogeneity at the single-cell level [26].
2.1.3. miRNAs
Circulating miRNAs, small 17- to 23-bp-long noncoding RNAs have been proposed as potential biomarkers for T2D [27].
In streptozotocin-treated mice, miRNA-375, which is highly abundant in pancreatic islet cells, was reported to be an early indicator of T1D [28].
miRNA-375, miRNA-101, and miRNA-802, which have roles in pancreatic islet function and insulin secretion, have been identified as being significantly increased in T2D patients [29].
Elevated levels of miR-122 were shown to be strongly associated with an increased risk of developing T2D [30]. MicroRNA-122 levels were also found to be increased in patients with diabetic retinopathy [31].
Large-scale studies are necessary for clinical miRNA validation. Since miRNAs are involved in diverse regulatory processes, future studies are needed to unravel their roles in the overall pathogenesis of T2D. A shift from targeted analysis towards exploratory next generation sequencing (NGS) should be performed in order to potentially discover novel miRNA relevant for T2D pathogenesis [32].
Circulating long-noncoding RNAs (lncRNAs), which are RNA molecules longer than 200 nucleotides and that do not code for proteins, exhibit cell and tissue specificity and have been implicated in various cellular processes, including transcriptional regulation. In the context of T2D complications, circulating lncRNAs have been shown to play important roles as transcriptional activators and regulators. In a mouse model of diabetic nephropathy, Tug1 lncRNA was shown to modulate mitochondrial bioenergetics in glomerular podocytes [33].
Other lncRNAs such as GAS5, RNA-MEG3is, MALAT1, and CYP4B1-PS1-001 were reported to be involved in diabetes-induced microvascular dysfunction and inflammation [34][35][36][37].
The use of lncRNAs comes with several disadvantages, however, including a lack of conservation across species and technically challenging analysis. Also, the fact that lncRNAs harbor a multitude of roles at the cellular level makes their specific functional characterization difficult.
Even though cfRNA provides a snapshot of transcripts reflecting the health status of multiple RNAs, understanding the precise origins of cfRNA, including their cell types of origin, can be challenging due to the complex and dynamic nature of cfRNA in circulation [38].
2.2. Extracellular Vesicles
EVs play a significant role in intercellular communication and their shedding is influenced by intracellular elements such as calcium ions. EVs can interact with recipient cells through various mechanisms, including ligand–receptor binding, phagocytosis, direct fusion with plasma membranes, micropinocytosis, and lipid raft-mediated internalization or endocytosis [39].
EVs are classified based on their different sizes and biogenesis processes (Figure 2). The main types of EVs are microparticles (MPs), exosomes, and apoptotic bodies, each with distinct characteristics [40]. MPs that are also known as microvesicles or ectosomes originate from the outward budding or shedding of the plasma membrane of cells and have a diameter size range from 30 nm to approximately 1 μm. Exosomes originate from the inward budding of endocytic vesicles within the cell, forming early endosomes which mature into late endosomes, also known as multivesicular bodies (MVBs), where intraluminal vesicles (ILVs) are formed through the inward budding of the endosomal membrane. Exosomes are released when MVBs fuse with the cell’s plasma membrane, leading to the extracellular release of the ILVs as exosomes. Exosomes are generally smaller, typically ranging from 30 to 150 nm in diameter. Apoptotic bodies are distinct from MPs and exosomes in terms of their origin and are typically larger than MPs and exosomes [41].
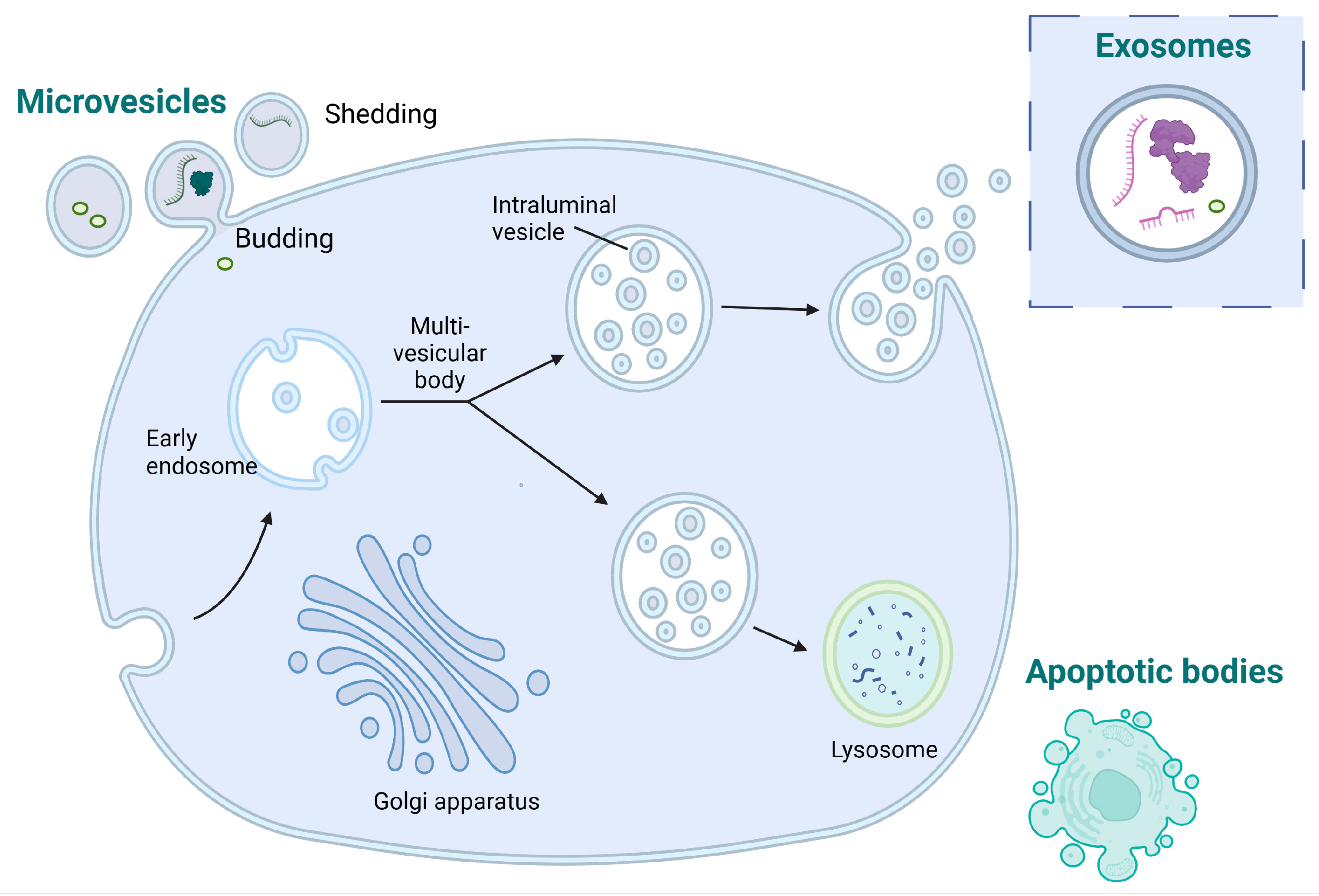
Figure 2. Diagram illustrating the processes involved in the generation of microvesicles, exosomes, and apoptotic bodies.
3. Salivary Markers
The conventional monitoring of serological parameters in diabetes typically involves invasive techniques, causing discomfort and distress. Collecting saliva is convenient, enhancing accessibility for both healthcare professionals and patients and is especially beneficial in scenarios where obtaining blood samples may pose challenges or discomfort. Saliva comprises a range of biomarkers, encompassing DNA, RNA, proteins (3000 proteins and 12,000 peptides), and metabolites [42]. An elegant review by Srinivasan et al. presented the wide array of salivary biomarkers linked to T2D, including glucose, glycosylated hemoglobin (HbA1c), amylase, alpha 2-macroglobulin, alpha defensins, carbonyls, dehydroprogesterone, ghrelin, high-density lipoprotein, C reactive protein, leptin, insulin, Il-1β, osteopontin, osteocalcin, transferrin receptor, urea, and uric acid [43][44].
In contrast to genetic T2D biomarkers, there is a limited body of research on mRNA biomarkers derived from saliva samples. A study examined differential gene expression in saliva from 13 T2D patients, revealing elevated levels of KRAS, SAT1, SLC13A2, and TMEM72, alongside reduced expressions of EGFR and PSMB2 genes [45]. Initially identified through microarray analysis, these six biomarkers were subsequently validated in a cohort of 13 T2D patients and 13 healthy controls. A logistic model incorporating four of these salivary biomarkers (KRAS, SAT1, EGFR, and PSMB2) successfully distinguished T2D patients from healthy controls, yielding an area under the curve (AUC) of 0.917 with a 95% confidence interval of 0.809–1.000. This suggests that salivary mRNAs hold promise as T2D biomarkers, although further investigations involving larger sample sizes are warranted. Subsequently, a validated panel comprising four salivary extracellular RNA biomarkers (IL1R2—interleukin 1 receptor type 2, VPS4B—vacuolar protein sorting 4 homolog B, CAP1—cyclase-associated actin cytoskeleton regulatory protein 1, LUZP6—leucine zipper protein 6) along with body mass index (BMI) has been identified. This panel demonstrates the capability to differentiate individuals with high and low insulin resistance, both in the general population and within subgroups of participants who are healthy and pre-diabetic [46].
There is still a need to identify reliable salivary biomarkers in order to diagnose prediabetes or to distinguish different T2D complications. For instance, differentiating between diabetic nephropathy (DN) and nondiabetic renal disease (NDRD) is crucial for tailoring more specific treatments. However, currently, there are no ideal biomarkers for making this distinction.
A recent study by Han et al. (2022) screened different salivary glycopatterns in DN and NDRD patients using lectin microarrays, with validation conducted through lectin blotting. Diagnostic models were then constructed using logistic regression and artificial neural network analyses, and their validity was confirmed in another cohort [47].
Oxidative stress occurs early in the development of T2D, often preceding the onset of clinical symptoms. Thus, monitoring oxidative stress markers allows for the detection of subtle changes at a molecular level before overt metabolic disturbances become evident [48]. Oxidative stress and inflammation can affect pancreatic beta cells, reducing their function and insulin secretion, further exacerbating glucose dysregulation. Prolonged exposure to oxidative stress, inflammation, and protein glycation contributes to the development of diabetic complications, including cardiovascular disease, neuropathy, nephropathy, and retinopathy. Understanding and targeting these interconnected pathways is essential for developing therapeutic interventions to manage T2D and prevent associated complications [49].
Numerous studies suggest that an imbalance in oxidant/antioxidant mediators plays a significant role in the development and advancement of metabolic syndrome, T2D, and cardiovascular disease. However, the majority of research has concentrated on the distribution of these indicators in tissues and blood, leaving their influence on saliva composition less explored. Products resulting from lipid peroxidation, protein oxidation, and DNA damage can be directly evaluated in saliva, potentially providing a means to diagnose systemic disorders associated with oxidative stress. The evaluation of salivary redox biomarkers appears to be applicable for both diagnosing and monitoring various health conditions, including obesity, diabetes, hypertensive disorders, and heart failure. In these conditions, there is a pathological depletion of molecules and enzymes with antioxidant properties in saliva, while oxidative and nitrosative by-products are favored. For instance, research has demonstrated elevated salivary oxidative biomarkers, including 4-hydroxynonenal (4-HNE), 8-isoprostanes (8-isoP), advanced oxidation protein products (AOPP), protein carbonyl groups (PC), and 8-hydroxy-D-guanosine (8-OHdG) [50].
The use of saliva samples for the molecular diagnostic of T2D comes with several caveats. For example, the local oral status and pathologies related to the oral cavity, such as periodontitis and dental caries may influence the redox balance of saliva, posing potential interference with its widespread routine clinical application [51]. Some biomarkers, such as pro-inflammatory cytokines, are common to many diseases, whereas others are more specific. The presence of other inflammatory comorbidities could also influence the reliability of salivary markers. Moreover, the oral microbiota, specific to each individual, might produce different metabolites and other signaling molecules which might hinder an accurate diagnostic.
The potential to identify a specific array of candidate molecules using saliva for distinguishing each stage of chronic disorder and creating panels of salivary mediators as appropriate molecular biomarkers, when integrated with the demographic, genetic, and anthropometric characteristics of patients, could offer an innovative diagnostic tool. Nevertheless, there is still a need to identify salivary markers specific for different T2D stages and complications. Once identified and validated in clinical studies, these biomarkers could further be incorporated into point-of-care devices or wearable sensors to enable their fast detection.
References
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas. Diabetes Res. Clin. Pract. 2019, 157, 107843.
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312.
- Iliescu, F.S.; Poenar, D.P.; Yu, F.; Ni, M.; Chan, K.H.; Cima, I.; Taylor, H.K.; Cima, I.; Iliescu, C. Recent advances in microfluidic methods in cancer liquid biopsy. Biomicrofluidics 2019, 13, 041503.
- Yaghoubi Naei, V.; Bordhan, P.; Mirakhorli, F.; Khorrami, M.; Shrestha, J.; Nazari, H.; Kulasinghe, A.; Ebrahimi Warkiani, M. Advances in novel strategies for isolation, characterization, and analysis of CTCs and ctDNA. Ther. Adv. Med. Oncol. 2023, 15, 17588359231192401.
- Armakolas, A.; Kotsari, M.; Koskinas, J. Liquid Biopsies, Novel Approaches and Future Directions. Cancers 2023, 15, 1579.
- Vaidyanathan, R.; Soon, R.H.; Zhang, P.; Jiang, K.; Lim, C.T. Cancer diagnosis: From tumor to liquid biopsy and beyond. Lab Chip 2019, 19, 11–34.
- Cima, I.; Wen Yee, C.; Iliescu, F.S.; Min Phyo, W.; Hon Lim, K.; Iliescu, C.; Han Tan, M. Label-free isolation of circulating tumor cells in microfluidic devices: Current research and perspectives. Biomicrofluidics 2013, 7, 011810.
- Arosemena, M.; Meah, F.A.; Mather, K.J.; Tersey, S.A.; Mirmira, R.G. Cell-free DNA fragments as biomarkers of islet β-cell death in obesity and type 2 diabetes. Int. J. Mol. Sci. 2021, 22, 2151.
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.-D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665.
- Khagi, Y.; Goodman, A.M.; Daniels, G.A.; Patel, S.P.; Sacco, A.G.; Randall, J.M.; Bazhenova, L.A.; Kurzrock, R. Hypermutated circulating tumor DNA: Correlation with response to checkpoint inhibitor–based immunotherapy. Clin. Cancer Res. 2017, 23, 5729–5736.
- Zhang, B.; Xu, C.W.; Shao, Y.; Wang, H.T.; Wu, Y.F.; Song, Y.Y.; Li, X.B.; Zhang, Z.; Wang, W.J.; Li, L.Q.; et al. Comparison of droplet digital PCR and conventional quantitative PCR for measuring EGFR gene mutation. Exp. Ther. Med. 2015, 9, 1383–1388.
- Imperial, R.; Nazer, M.; Ahmed, Z.; Kam, A.E.; Pluard, T.J.; Bahaj, W.; Levy, M.; Kuzel, T.M.; Hayden, D.M.; Pappas, S.G.; et al. Matched whole-genome sequencing (WGS) and whole-exome sequencing (WES) of tumor tissue with circulating tumor DNA (ctDNA) analysis: Complementary modalities in clinical practice. Cancers 2019, 11, 1399.
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.; Kaper, F.; Dawson, S.-J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012, 4, 136ra168.
- Akirav, E.M.; Lebastchi, J.; Galvan, E.M.; Henegariu, O.; Akirav, M.; Ablamunits, V.; Lizardi, P.M.; Herold, K.C. Detection of β cell death in diabetes using differentially methylated circulating DNA. Proc. Natl. Acad. Sci. USA 2011, 108, 19018–19023.
- El Tarhouny, S.A.; Hadhoud, K.M.; Ebrahem, M.M.; Al Azizi, N.M. Assessment of cell-free DNA with microvascular complication of type II diabetes mellitus, using PCR and ELISA. Nucleosides Nucleotides Nucleic Acids 2010, 29, 228–236.
- Peng, X.; Li, H.-D.; Wu, F.-X.; Wang, J. Identifying the tissues-of-origin of circulating cell-free DNAs is a promising way in noninvasive diagnostics. Brief. Bioinform. 2021, 22, bbaa060.
- Nishimoto, S.; Fukuda, D.; Higashikuni, Y.; Tanaka, K.; Hirata, Y.; Murata, C.; Kim-Kaneyama, J.-R.; Sato, F.; Bando, M.; Yagi, S. Obesity-induced DNA released from adipocytes stimulates chronic adipose tissue inflammation and insulin resistance. Sci. Adv. 2016, 2, e1501332.
- Lehmann-Werman, R.; Neiman, D.; Zemmour, H.; Moss, J.; Magenheim, J.; Vaknin-Dembinsky, A.; Rubertsson, S.; Nellgård, B.; Blennow, K.; Zetterberg, H. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl. Acad. Sci. USA 2016, 113, E1826–E1834.
- Karaglani, M.; Panagopoulou, M.; Cheimonidi, C.; Tsamardinos, I.; Maltezos, E.; Papanas, N.; Papazoglou, D.; Mastorakos, G.; Chatzaki, E. Liquid Biopsy in Type 2 Diabetes Mellitus Management: Building Specific Biosignatures via Machine Learning. J. Clin. Med. 2022, 11, 1045.
- Birinci, A.; Birinci, H.; Abidinoglu, R.; Durupinar, B.; Oge, I. Diabetic retinopathy and HLA antigens in type 2 diabetes mellitus. Eur. J. Ophthalmol. 2002, 12, 89–93.
- Ma, Z.J.; Sun, P.; Guo, G.; Zhang, R.; Chen, L.M. Association of the HLA-DQA1 and HLA-DQB1 Alleles in Type 2 Diabetes Mellitus and Diabetic Nephropathy in the Han Ethnicity of China. J. Diabetes Res. 2013, 2013, 452537.
- Trumpff, C.; Michelson, J.; Lagranha, C.J.; Taleon, V.; Karan, K.R.; Sturm, G.; Lindqvist, D.; Fernström, J.; Moser, D.; Kaufman, B.A.; et al. Stress and circulating cell-free mitochondrial DNA: A systematic review of human studies, physiological considerations, and technical recommendations. Mitochondrion 2021, 59, 225–245.
- Liu, J.; Zou, Y.; Tang, Y.; Xi, M.; Xie, L.; Zhang, Q.; Gong, J. Circulating cell-free mitochondrial deoxyribonucleic acid is increased in coronary heart disease patients with diabetes mellitus. J. Diabetes Investig. 2016, 7, 109–114.
- Silzer, T.; Barber, R.; Sun, J.; Pathak, G.; Johnson, L.; O’Bryant, S.; Phillips, N. Circulating mitochondrial DNA: New indices of type 2 diabetes-related cognitive impairment in Mexican Americans. PLoS ONE 2019, 14, e0213527.
- Umu, S.U.; Langseth, H.; Bucher-Johannessen, C.; Fromm, B.; Keller, A.; Meese, E.; Lauritzen, M.; Leithaug, M.; Lyle, R.; Rounge, T.B. A comprehensive profile of circulating RNAs in human serum. RNA Biol. 2018, 15, 242–250.
- Guruprasad, P.; Lee, Y.G.; Kim, K.H.; Ruella, M. The current landscape of single-cell transcriptomics for cancer immunotherapy. J. Exp. Med. 2020, 218, e20201574.
- Alvarez, M.L.; DiStefano, J.K. The role of non-coding RNAs in diabetic nephropathy: Potential applications as biomarkers for disease development and progression. Diabetes Res. Clin. Pract. 2013, 99, 1–11.
- Erener, S.; Mojibian, M.; Fox, J.K.; Denroche, H.C.; Kieffer, T.J. Circulating miR-375 as a biomarker of β-cell death and diabetes in mice. Endocrinology 2013, 154, 603–608.
- Higuchi, C.; Nakatsuka, A.; Eguchi, J.; Teshigawara, S.; Kanzaki, M.; Katayama, A.; Yamaguchi, S.; Takahashi, N.; Murakami, K.; Ogawa, D. Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism 2015, 64, 489–497.
- Willeit, P.; Skroblin, P.; Moschen, A.R.; Yin, X.; Kaudewitz, D.; Zampetaki, A.; Barwari, T.; Whitehead, M.; Ramírez, C.M.; Goedeke, L. Circulating microRNA-122 is associated with the risk of new-onset metabolic syndrome and type 2 diabetes. Diabetes 2017, 66, 347–357.
- Pastukh, N.; Meerson, A.; Kalish, D.; Jabaly, H.; Blum, A. Serum miR-122 levels correlate with diabetic retinopathy. Clin. Exp. Med. 2019, 19, 255–260.
- Turchinovich, A.; Baranova, A.; Drapkina, O.; Tonevitsky, A. Cell-free circulating nucleic acids as early biomarkers for NAFLD and NAFLD-associated disorders. Front. Physiol. 2018, 9, 1256.
- Long, J.; Badal, S.S.; Ye, Z.; Wang, Y.; Ayanga, B.A.; Galvan, D.L.; Green, N.H.; Chang, B.H.; Overbeek, P.A.; Danesh, F.R. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J. Clin. Investig. 2016, 126, 4205–4218.
- Carter, G.; Miladinovic, B.; Patel, A.A.; Deland, L.; Mastorides, S.; Patel, N.A. Circulating long noncoding RNA GAS5 levels are correlated to prevalence of type 2 diabetes mellitus. BBA Clin. 2015, 4, 102–107.
- Qiu, G.-Z.; Tian, W.; Fu, H.-T.; Li, C.-P.; Liu, B. Long noncoding RNA-MEG3 is involved in diabetes mellitus-related microvascular dysfunction. Biochem. Biophys. Res. Commun. 2016, 471, 135–141.
- Li, X.; Zeng, L.; Cao, C.; Lu, C.; Lian, W.; Han, J.; Zhang, X.; Zhang, J.; Tang, T.; Li, M. Long noncoding RNA MALAT1 regulates renal tubular epithelial pyroptosis by modulated miR-23c targeting of ELAVL1 in diabetic nephropathy. Exp. Cell Res. 2017, 350, 327–335.
- Wang, M.; Wang, S.; Yao, D.; Yan, Q.; Lu, W. A novel long non-coding RNA CYP4B1-PS1-001 regulates proliferation and fibrosis in diabetic nephropathy. Mol. Cell. Endocrinol. 2016, 426, 136–145.
- Chalasani, N.; Toden, S.; Sninsky, J.J.; Rava, R.P.; Braun, J.V.; Gawrieh, S.; Zhuang, J.; Nerenberg, M.; Quake, S.R.; Maddala, T. Noninvasive stratification of nonalcoholic fatty liver disease by whole transcriptome cell-free mRNA characterization. Am. J. Physiol.-Gastrointest. Liver Physiol. 2021, 320, G439–G449.
- Fierabracci, A.; Del Fattore, A.; Muraca, M.; Vittorio Delfino, D.; Muraca, M. The use of mesenchymal stem cells for the treatment of autoimmunity: From animals models to human disease. Curr. Drug Targets 2016, 17, 229–238.
- Rupert, D.L.; Claudio, V.; Lässer, C.; Bally, M. Methods for the physical characterization and quantification of extracellular vesicles in biological samples. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2017, 1861, 3164–3179.
- La Marca, V.; Fierabracci, A. Insights into the diagnostic potential of extracellular vesicles and their miRNA signature from liquid biopsy as early biomarkers of diabetic micro/macrovascular complications. Int. J. Mol. Sci. 2017, 18, 1974.
- Loo, J.A.; Yan, W.; Ramachandran, P.; Wong, D.T. Comparative human salivary and plasma proteomes. J. Dent. Res. 2010, 89, 1016–1023.
- Srinivasan, M.; Blackburn, C.; Mohamed, M.; Sivagami, A.V.; Blum, J. Literature-based discovery of salivary biomarkers for type 2 diabetes mellitus. Biomark Insights 2015, 10, 39–45.
- Marques, R.C.R.; da Silva, J.R.; Lima, C.P.V.; Stefani, C.M.; Damé-Teixeira, N. Salivary parameters of adults with diabetes mellitus: A systematic review and meta-analysis. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2022, 134, 176–189.
- Lee, Y.-H.; Joshipura, K.; Vergara, J.L.; Wong, D.T. Detection of Type II Diabetes Mellitus Using Salivary Transcriptomic Biomarkers. Genom. Med. Biomark. Health Sci. 2012, 4, 7–11.
- Zhang, Y.; Sun, J.; Li, F.; Grogan, T.R.; Vergara, J.L.; Luan, Q.; Park, M.S.; Chia, D.; Elashoff, D.; Joshipura, K.J.; et al. Salivary extracellular RNA biomarkers for insulin resistance detection in hispanics. Diabetes Res. Clin. Pract. 2017, 132, 85–94.
- Han, Q.; Wang, X.; Ding, X.; Hao, J.; Li, Q.; Wang, J.; Yu, H.; Tang, Z.; Yang, F.; Cai, G.; et al. Salivary Glycopatterns as Potential Non-Invasive Biomarkers for Diagnosing and Reflecting Severity and Prognosis of Diabetic Nephropathy. Front. Endocrinol. 2022, 13, 790586.
- Anwar, S.; Khan, M.A.; Sadaf, A.; Younus, H. A structural study on the protection of glycation of superoxide dismutase by thymoquinone. Int. J. Biol. Macromol. 2014, 69, 476–481.
- Rahmani, A.H.; Anwar, S.; Raut, R.; Almatroudi, A.; Babiker, A.Y.; Khan, A.A.; Alsahli, M.A.; Almatroodi, S.A. Therapeutic Potential of Myrrh, a Natural Resin, in Health Management through Modulation of Oxidative Stress, Inflammation, and Advanced Glycation End Products Formation Using In Vitro and In Silico Analysis. Appl. Sci. 2022, 12, 9175.
- Fejfer, K.; Buczko, P.; Niczyporuk, M.; Ładny, J.R.; Hady, H.R.; Knaś, M.; Waszkiel, D.; Klimiuk, A.; Zalewska, A.; Maciejczyk, M. Oxidative modification of biomolecules in the nonstimulated and stimulated saliva of patients with morbid obesity treated with bariatric surgery. Biomed. Res. Int. 2017, 2017, 4923769.
- Tóthová, L.U.; Kamodyova, N.; Červenka, T.; Celec, P. Salivary markers of oxidative stress in oral diseases. Front. Cell Infect. Microbiol. 2015, 5, 73.
More
Information
Subjects:
Medicine, Research & Experimental
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
412
Revisions:
2 times
(View History)
Update Date:
06 Mar 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




