Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Carmine Crecchio | -- | 1521 | 2024-03-04 10:54:15 | | | |
| 2 | Rita Xu | Meta information modification | 1521 | 2024-03-04 11:02:22 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Khanghahi, M.Y.; Strafella, S.; Filannino, P.; Minervini, F.; Crecchio, C. Lactic Acid Bacteria as an Emerging Group. Encyclopedia. Available online: https://encyclopedia.pub/entry/55818 (accessed on 07 February 2026).
Khanghahi MY, Strafella S, Filannino P, Minervini F, Crecchio C. Lactic Acid Bacteria as an Emerging Group. Encyclopedia. Available at: https://encyclopedia.pub/entry/55818. Accessed February 07, 2026.
Khanghahi, Mohammad Yaghoubi, Sabrina Strafella, Pasquale Filannino, Fabio Minervini, Carmine Crecchio. "Lactic Acid Bacteria as an Emerging Group" Encyclopedia, https://encyclopedia.pub/entry/55818 (accessed February 07, 2026).
Khanghahi, M.Y., Strafella, S., Filannino, P., Minervini, F., & Crecchio, C. (2024, March 04). Lactic Acid Bacteria as an Emerging Group. In Encyclopedia. https://encyclopedia.pub/entry/55818
Khanghahi, Mohammad Yaghoubi, et al. "Lactic Acid Bacteria as an Emerging Group." Encyclopedia. Web. 04 March, 2024.
Copy Citation
Increasing awareness of the problems caused by synthetic agrochemicals, such as chemical fertilizers, pesticides, and herbicides, makes it crucial to discover substitute approaches that can guarantee competitive plant production and protect the environment while maintaining the natural balance in agroecosystems. One of the leading alternatives is utilizing rhizobacterial strains named plant growth-promoting rhizobacteria (PGPR).
biofertilization
bioprotection
plant growth-promoting rhizobacteria
1. Introduction
Supplying adequate agrifood products and byproducts, the demand for which has increased as the global population rises, requires diverse strategies, including (i) incrementing the cultivation area and (ii) improving the production per unit area. However, although the first approach (e.g., land use change) increased production, the conversion of natural landforms into farming land led to an environmental challenge through land degradation [1][2][3]. This problem has become increasingly important in the Mediterranean basin, which demonstrates obvious movements of degradation, especially in areas where climate changes and meteorological conditions contribute extremely to it [4]. As an alternative strategy, the application of agrochemicals (e.g., artificial fertilizers, herbicides, etc.) and intensive farming management practices to increase crop production has brought the major disadvantage of the increasing contamination of agricultural products and the environment [5][6]. Moreover, the strong demand for agrochemicals from domestic and global markets has driven up their prices and caused economic challenges in the agricultural sector [7].
One of the most significant current discussions is a reconsideration of technologies to boost plant production, focusing on alternative strategies, mainly the application of beneficial biological approaches and bio-based products. The application of beneficial microbial consortia, mainly plant growth-promoting rhizobacteria (PGPR), has become one of the most widely used biological alternatives in sustainable agriculture. Such beneficial rhizobacteria can be considered plant biostimulants, which, according to Regulation (EU) No. 2019/1009 of the European Parliament [8], can be used as fertilizing products to promote the plant nutritional value, increase the nutrient locked-up availability in the rhizosphere/soil, and improve plant tolerance against abiotic and biotic stresses [9]. Moreover, the presented sustainable alternatives of agrochemicals receive considerable critical attention in fulfilling part of the United Nations Sustainable Development Goal 15, including how microbial-based biofertilization can promote the sustainable use of agroecosystems and preserve farmlands from degradation.
Recently, investigators have attempted to evaluate the potential of identified PGPR and their mechanisms of action as agents of biofertilization, phytostimulation, bioremediation, and bioprotection (Figure 1).
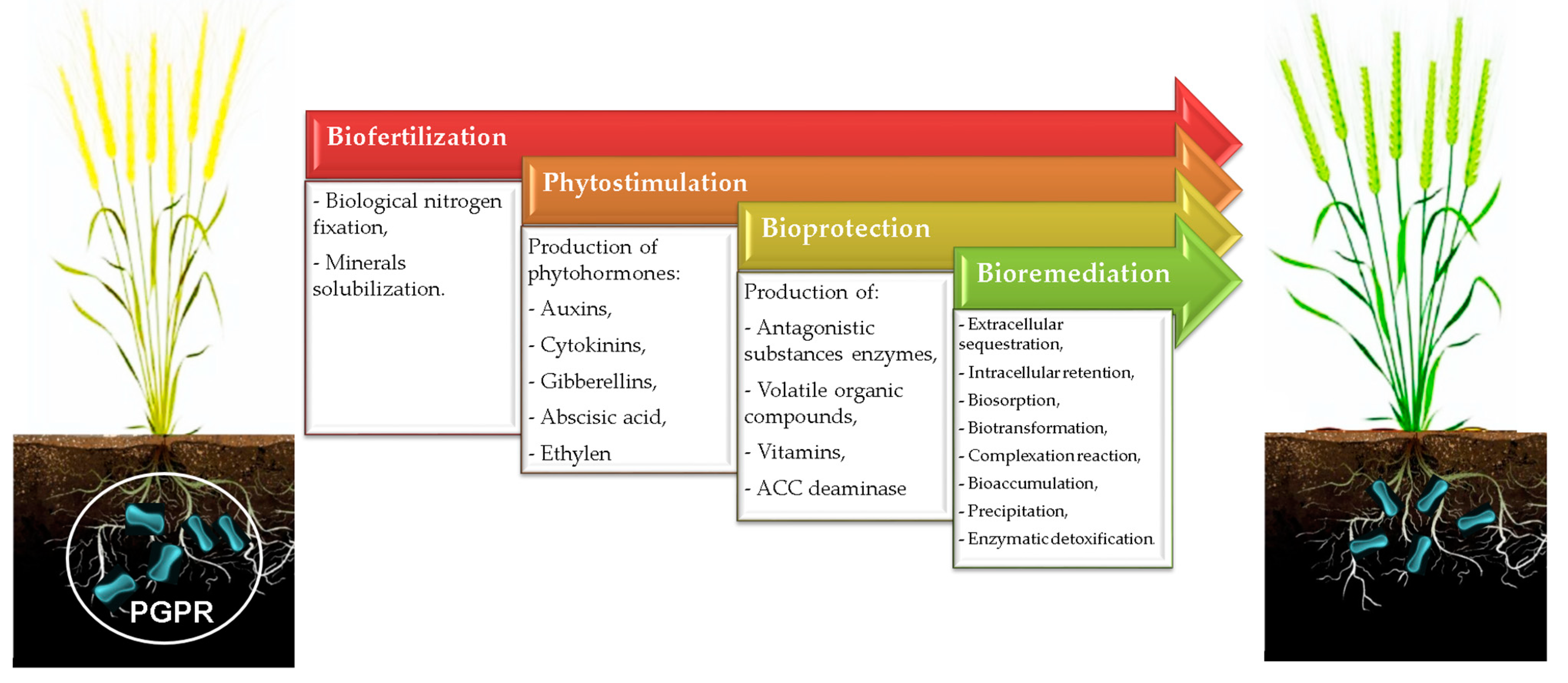
Figure 1. Potential roles of PGPR in agroecosystems.
2. Lactic Acid Bacteria (LAB): An Emerging Group of PGPR
2.1. Soil- and Plant-Associated LAB
Diverse genera of beneficial rhizobacteria have already been proposed as PGPR, with Bacillus and Pseudomonas being the predominant genera [10]. Nevertheless, metagenomic analyses of plant and rhizosphere microbiomes have resulted in the identification of an emerging group of PGPR, namely, lactic acid bacteria (LAB), which are barely detectable in the plant–soil ecosystem due to their low abundance [11][12]. LAB are known as microaerophilic, Gram-positive, cytochrome-deficient, and nonsporulating bacteria that are also involved in food and silage fermentation as well as soil health; however, some of them are recognized as human pathogens [13][14]. Despite intensive investigations into the conventional function of LAB in the food processing industry, too little attention has been given to their other functions, such as acting as biofertilizers, biocontrol agents, and biostimulants in plant growth. Furthermore, little is known about LAB due to the difficulty of isolating them by plating serial dilutions of rhizospheric soil samples since enrichment methods using selective culture media have been largely ignored [11][15][16]. Despite their low relative abundances, LAB have been isolated from the rhizosphere in some studies, which has consequently led to their introduction as a crucial component of sustainable agricultural approaches as environmentally sustainable and efficient strategies to control pests and diseases and enhance crop yield [16][17][18].
It has been stated that root exudates, including amino acids, carbohydrates, enzymes, organic acids, phenols, and flavonoids, account for a considerable proportion (5–21%) of photosynthetically fixed carbon in plants, which can change the soil environment and, consequently, shape microbial communities [19]. Although such a carbohydrate-rich rhizosphere is ideal for LAB, a quick breakdown of organic acids in the rhizosphere has been proposed as a limiting factor in the capability of LAB to acidify soil to their benefit, thus preventing LAB from being the predominant taxon in agricultural soils [20]. Moreover, recent research on the efficient transfer of LAB from the rhizosphere and phyllosphere to the plant endosphere [21] provided an interesting strategy to assess their roles in plant growth and production. In fact, LAB also constitute a small fraction of the epiphyte [22][23] and endophyte populations of plant microbiota [23][24]. Among the plant-associated LAB, there are some well-known generalist taxa, including Lactiplantibacillus plantarum, Lactococcus lactis, Leuconostoc spp., Weissella spp., and Enterococcus spp., and some specialist taxa, such as Fructilactobacillus florum and Fructobacillus spp., that have been discovered relatively recently [25]. However, the consequences of LAB on plant physiology still need to be fully deciphered. Overall, the genomic diversity in LAB is mainly due to the particular pressure applied by each plant niche [26][27].
2.2. Biofertilization and Bioremediation Effects of LAB
The ability of LAB to synthesize metabolites, including organic acids, phenolic acids and their flavonoid derivatives, phytohormones, and antimicrobial substrates, has already been reported [28][29][30][31]. LAB have been reported to have a high capacity to solubilize insoluble forms of phosphate [28][32][33] and potassium [11], to biologically fix nitrogen [34], and to produce iron-chelating compounds [35] and siderophores [36]. They are also involved in soil biochemical cycles through regulating soil organic matter content and detoxifying hazardous chemicals [35]. Heavy metal biosorption mechanisms of LAB have been previously reported, involving bacterial surface-associated functional groups, including carboxyl, hydroxyl, and phosphate [35][37]. Previous studies on food technology outlined the critical role of LAB in breaking down organic macromolecules and indigestible polysaccharides and converting disfavored flavor compounds [38].
In addition, it has already been suggested that shifts in the microbiome in response to environmental changes may imply the plasticity of the available microbial genetic pool in aiding plant adaptation to environmental stress [39][40]. Accordingly, the finding of a rich diversity of LAB in the rhizosphere of plants grown in deserts [16][20] can confirm the role of LAB in improving the tolerance of associated plants. It can also be assumed that LAB conferring a specific stress tolerance can be derived from holobionts thriving under similar stress conditions. Improved tolerance of LAB-treated plants to abiotic stresses has been correlated with changes in plant metabolic responses related to proline content, phenolic acids, and antioxidant enzymes [20][41]. Such reported findings can support the assumption that LAB are effective as biofertilizers by increasing nutrient bioavailability and as biostimulants to stimulate plant growth or seed germination by alleviating diverse environmental stresses [20][42][43]. A scheme of the biofertilization, bioprotection, and biodegradation potential of LAB is shown in Figure 2.

Figure 2. Schematic portrayal of the biofertilization, bioprotection, and biodegradation potential of lactic acid bacteria.
2.3. Bioprotection Effects of LAB
LAB have also received considerable attention for their capability to synthesize antifungal metabolites (e.g., diketopiperazines, hydroxy derivatives of unsaturated fatty acids, and 3-phenyllactic acid), antibacterial (e.g., bacteriocins and bacteriocin-like substances), and general antimicrobial metabolites (e.g., hydrogen peroxide, organic acids, pyrrolidone-5-carboxylic acid, diacetyl, and reuterin) [44][45][46]. In addition to direct antagonism against pathogens, LAB can affect the plant response to pathogens by causing systemic acquired resistance (SAR) and enhancing plant innate immunity [20]. Mao et al. [47] observed that the antibacterial activity of Lacti plantarum DY-6 was dependent on the production of acetic acid, lactic acid, caprylic acid, propionic acid, and decyl acid. On the other hand, Magnusson et al. [48] observed that the ability to synthesize lactic acid in the bacterial strains without antimicrobial activity was in the same range or even higher than those possessing antimicrobial activity, while the amount of acetic acid corresponded to that normally detected in the culture medium used for assessment tests. Therefore, they concluded that the antimicrobial activity of Lacti. plantarum, Latilactobacillus sakei, Loigolactobacillus coryniformis, and Pediococcus. pentosaceus against Aspergillus sp., Fusarium sp., and Penicillium sp. was due to the synthesis of other metabolites [48]. Through an HPLC analysis of antagonistic bacteria supernatants, they detected two antifungal cyclic dipeptides, cyclo (Phe-Pro) and cyclo (Phe-4-OH-Pro), whose structures were similar to those found in Lacti. plantarum by Ström et al. [49]. Axel et al. [50] found that chemical acidification has no effect on mold inhibition in food, so it is more plausible that the antagonistic activity of LAB depends on the synergistic action between organic acids and other active compounds [51]. The production of bacteriocins by soil- and plant-associated LAB is rare but not excluded, as it was observed that the treatments of cell-free supernatants with organic solvents, surfactants, H2O2, high temperature, and different pH do not affect their antimicrobial activity [52]. Yanagida et al. [53] were the first to report the production of bacteriocins by Ligilactobacillus animalis C060203 and Enterococcus durans C102901, which exhibited strong antibacterial activity against Lati. sakei JCM 1157T. The defeat of antibacterial potential in response to proteinase K treatment confirmed the proteinaceous nature of antimicrobial compounds. A comparative genomic analysis between LAB isolated from plant/soil ecosystems and those isolated from dairy products, nondomestic animals, and human isolates revealed that plant/soil LAB are enriched in genes involved in bacteriocin synthesis, suggesting a probable role in plant fitness [11]. This finding confirms that LAB are a natural farm of antimicrobial metabolites [44] and can be used in agronomic fields to prevent or relieve disease sustainably.
References
- Cerdà, A.; Hooke, J.; Romero-Diaz, A.; Montanarella, L.; Lavee, H. Soilerosion on Mediterraneantype-ecosystems. Land Degrad Dev. 2010, 21, 71–74.
- Yaghoubi Khanghahi, M.; Murgese, P.; Strafella, S.; Crecchio, C. Soil Biological Fertility and Bacterial Community Response to Land Use Intensity: A Case Study in the Mediterranean Area. Diversity 2019, 11, 211.
- Yaghoubi Khanghahi, M.; Cucci, G.; Lacolla, G.; Lanzellotti, L.; Crecchio, C. Soil fertility and bacterial community composition in a semiarid Mediterranean agricultural soil under long-term tillage management. Soil Use Manage 2020, 36, 604–615.
- Racioppo, A.; d’Amelio, A.; De Santis, A.; Bevilacqua, A.; Corbo, M.R.; Sinigaglia, M. Potential Use of Plant Growth-Promoting Bacteria to Enhance Growth and Soil Fertility in Marginal Areas: Focus on the Apulia Region, Italy. Agronomy 2023, 13, 2983.
- Meena, V.S.; Meena, S.K.; Verma, J.P.; Kumar, A.; Aeron, A.; Mishra, P.K.; Bisht, J.K.; Pattanayak, A.; Naveed, M.; Dotaniya, M.L. Plant beneficial rhizospheric microorganism (PBRM) strategies to improve nutrients use efficiency: A review. Ecol. Eng. 2017, 107, 8–32.
- Yaghoubi Khanghahi, M.; Pirdashti, H.; Rahimian, H.; Nematzadeh, G.A.; Ghajar Sepanlou, M. Nutrient use efficiency and nutrient uptake promoting of rice by potassium solubilizing bacteria (KSB). Cereal Res. Commun. 2018, 46, 739–750.
- Nishimoto, R. Global trends in the crop protection industry. J. Pestic. Sci. 2019, 44, 141–147.
- EUR-Lex—32019R1009—EN—EUR-Lex. Available online: https://eur-lex.europa.eu/eli/reg/2019/1009/oj (accessed on 10 December 2023).
- Hendriksen, N.B. Microbial Biostimulants—The Need for Clarification in EU Regulation. Trends Microbiol. 2022, 30, 311–313.
- Beneduzi, A.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 2012, 35, 1044–1051.
- Strafella, S.; Simpson, D.J.; Yaghoubi Khanghahi, M.; De Angelis, M.; Gänzle, M.; Minervini, F.; Crecchio, C. Comparative Genomics and In Vitro Plant Growth Promotion and Biocontrol Traits of Lactic Acid Bacteria from the Wheat Rhizosphere. Microorganisms 2021, 9, 78.
- Jaffar, N.S.; Jawan, R.; Chong, K.P. The potential of lactic acid bacteria in mediating the control of plant diseases and plant growth stimulation in crop production—A mini review. Front. Plant Sci. 2023, 13, 1047945.
- Duar, R.M.; Lin, X.B.; Zheng, J.; Martino, M.E.; Grenier, T.; Pérez-Muñoz, M.E.; Leulier, F.; Gänzle, M.; Walter, J. Lifestyles in transition: Evolution and natural history of the genus Lactobacillus. FEMS Microbiol. Rev. 2017, 41, S27–S48.
- Bintsis, T. Lactic acid bacteria as starter cultures: An update in their metabolism and genetics. AIMS Microbiol. 2018, 4, 665–684.
- Chen, Y.S.; Yanagida, F.; Shinohara, T. Isolation and identification of lactic acid bacteria from soil using an enrichment procedure. Lett. Appl. Microbial. 2005, 40, 195–200.
- Fhoula, I.; Najjari, A.; Turki, Y.; Jaballah, S.; Boudabous, A.; Ouzari, H. Diversity and antimicrobial properties of lactic acid bacteria isolated from rhizosphere of olive trees and desert truffles of Tunisia. BioMed Res. Int. 2013, 2013, 405708.
- Lutz, M.P.; Michel, V.; Martinez, C.; Camps, C. Lactic acid bacteria as biocontrol agents of soil-borne pathogens. Bull. IOBC/WPRS 2012, 78, 285–288.
- Murindangabo, Y.T.; Kopecký, M.; Perná, K.; Nguyen, T.G.; Konvalina, P.; Kavková, M. Prominent use of lactic acid bacteria in soil-plant systems. Appl. Soil Ecol. 2023, 189, 104955.
- Liao, Z.; Fan, J.; Lai, Z.; Bai, Z.; Wang, H.; Cheng, M.; Zhang, F.; Li, Z. Chapter Three—Response network and regulatory measures of plant-soil-rhizosphere environment to drought stress. Adv. Agron. 2023, 180, 93–196.
- Lamont, J.R.; Wilkins, O.; Bywater-Ekegärd, M.; Smith, D.L. From yogurt to yield: Potential applications of lactic acid bacteria in plant production. Soil Biol. Biochem. 2017, 111, 1–9.
- Viscardi, S.; Marileo, L.; Barra, P.J.; Durán, P.; Inostroza-Blancheteau, C. From farm to fork: It could be the case of Lactic Acid Bacteria in the stimulation of folates biofortification in food crops. Curr. Opin. Food Sci. 2020, 34, 1–8.
- Di Cagno, R.; Coda, R.; De Angelis, M.; Gobbetti, M. Exploitation of vegetables and fruits through lactic acid fermentation. Food Microbiol. 2013, 33, 1–10.
- Pontonio, E.; Di Cagno, R.; Tarraf, W.; Filannino, P.; De Mastro, G.; Gobbetti, M. Dynamic and assembly of epiphyte and endophyte lactic acid bacteria during the life cycle of Origanum vulgare L. Front. Microbiol. 2018, 9, 1372.
- Minervini, F.; Celano, G.; Lattanzi, A.; Tedone, L.; De Mastro, G.; Gobbetti, M.; De Angelis, M. Lactic acid bacteria in durum wheat flour are endophytic components of the plant during its entire life cycle. Appl. Environ. Microbiol. 2015, 81, 6736–6748.
- Yu, A.O.; Leveau, J.H.; Marco, M.L. Abundance, diversity and plant-specific adaptations of plant-associated lactic acid bacteria. Environ. Microbiol. Rep. 2020, 12, 16–29.
- McAuliffe, O. Symposium review: Lactococcus lactis from nondairy sources: Their genetic and metabolic diversity and potential applications in cheese1. J. Dairy Sci. 2018, 101, 3597–3610.
- Golomb, B.L.; Marco, M.L. Lactococcus lactis metabolism and gene expression during growth on plant tissues. J. Bacteriol. 2015, 197, 371.
- Murgese, P.; Santamaria, P.; Leoni, B.; Crecchio, C. Ameliorative effects of PGPB on yield, physiological parameters, and nutrient transporter genes expression in barattiere (Cucumis melo L.). J. Soil Sci. Plant Nutr. 2020, 20, 784–793.
- Dalié, D.K.D.; Deschamps, A.M.; Richard-Forget, F. Lactic acid bacteria—Potential for control of mould growth and mycotoxins: A review. Food Control. 2010, 21, 370–380.
- Filannino, P.; Gobbetti, M.; De Angelis, M.; Di Cagno, R. Hydroxycinnamic acids used as external acceptors of electrons: An energetic advantage for strictly heterofermentative lactic acid bacteria. Appl. Environ. Microbiol. 2014, 80, 7574–7582.
- Filannino, P.; Di Cagno, R.; Gobbetti, M. Metabolic and functional paths of lactic acid bacteria in plant foods: Get out of the labyrinth. Curr. Opin. Biotechnol. 2018, 49, 64–72.
- De Lacerda, J.R.M.; Da Silva, T.F.; Vollú, R.E.; Marques, J.M.; Seldin, L. Generally recognized as safe (GRAS) Lactococcus lactis strains associated with Lippia sidoides Cham. are able to solubilize/mineralize phosphate. Springer Plus 2016, 5, 828.
- Mussa, A.; Million, T.; Assefa, F. Rhizospheric bacterial isolates of grass pea (Lathyrus sativus L.) endowed with multiple plant growth promoting traits. J. Appl. Microbiol. 2018, 125, 1786–1801.
- Giassi, V.; Kiritani, C.; Kupper, K.C. Bacteria as growth-promoting agents for citrus rootstocks. Microbiol. Res. 2016, 190, 46–54.
- Raman, J.; Kim, J.-S.; Choi, K.R.; Eun, H.; Yang, D.; Ko, Y.-J.; Kim, S.-J. Application of Lactic Acid Bacteria (LAB) in Sustainable Agriculture: Advantages and Limitations. Int. J. Mol. Sci. 2022, 23, 7784.
- Shrestha, A.; Kim, B.S.; Park, D.H. Biological control of bacterial spot disease and plant growth-promoting effects of lactic acid bacteria on pepper. Biocontrol Sci. Technol. 2014, 24, 763–779.
- Ameen, F.A.; Hamdan, A.M.; El-Naggar, M.Y. Assessment of the heavy metal bioremediation efficiency of the novel marine lactic acid bacterium, Lactobacillus plantarum MF042018. Sci. Rep. 2020, 10, 314.
- Wang, Y.; Wu, J.; Lv, M.; Shao, Z.; Hungwe, M.; Wang, J.; Bai, X.; Xie, J.; Wang, Y.; Geng, W. Metabolism Characteristics of Lactic Acid Bacteria and the Expanding Applications in Food Industry. Front. Bioeng. Biotechnol. 2021, 9, 612285.
- Mukherjee, A.; Singh, S.; Gaurav, A.K.; Chouhan, G.K.; Jaiswal, D.K.; de Araujo Pereira, A.P.; Passari, A.K.; Abdel-Azeem, A.M.; Verma, J.P. Harnessing of phytomicrobiome for developing potential biostimulant consortium for enhancing the productivity of chickpea and soil health under sustainable agriculture. Sci. Total Environ. 2022, 836, 155550.
- Bandopadhyay, S.; Li, X.; Bowsher, A.W.; Last, R.L.; Shade, A. Disentangling plant- and environment-mediated drivers of active rhizosphere bacterial community dynamics during short-term drought. BioRxiv 2023, preprint.
- Phoboo, S.; Sarkar, D.; Bhowmik, P.C.; Jha, P.K.; Shetty, K. Improving salinity resilience in Swertia chirayita clonal line with Lactobacillus plantarum. Can. J. Plant Sci. 2016, 96, 117–127.
- Daranas, N.; Roselló, G.; Cabrefiga, J.; Donati, I.; Francés, J.; Badosa, E.; Spinelli, F.; Montesinos, E.; Bonaterra, A. Biological control of bacterial plant diseases with Lactobacillus plantarum strains selected for their broad-spectrum activity. Ann. Appl. Biol. 2019, 174, 92–105.
- Afanador-Barajas, L.N.; Navarro-Noya, Y.E.; Luna-Guido, M.L. Impact of a bacterial consortium on the soil bacterial community structure and maize (Zea mays L.) cultivation. Sci. Rep. 2021, 11, 13092.
- Stoyanova, L.G.; Ustiugova, E.A.; Netrusov, A.I. Antibacterial metabolites of lactic acid bacteria: Their diversity and properties. Prikl. Biokhim. Mikrobiol. 2012, 48, 259–275.
- Benítez-Chao, D.F.; León-Buitimea, A.; Lerma-Escalera, J.A.; Morones-Ramírez, J.R. Bacteriocins: An Overview of Antimicrobial, Toxicity, and Biosafety Assessment by in vivo Models. Front. Microbiol. 2021, 12, 630695.
- Fiorino, G.M.; Tlais, A.Z.A.; Losito, I.; Filannino, P.; Gobbetti, M.; Di Cagno, R. Triacylglycerols hydrolysis and hydroxy-and epoxy-fatty acids release during lactic fermentation of plant matrices: An extensive study showing inter-and intra-species capabilities of lactic acid bacteria. Food Chem. 2023, 412, 135552.
- Mao, Y.; Zhang, X.; Xu, Z. Identification of Antibacterial Substances of Lactobacillus plantarum DY-6 for Bacteriostatic Action. Food Sci. Nutr. 2020, 8, 2854–2863.
- Magnusson, J.; Ström, K.; Roos, S.; Sjögren, J.; Schnürer, J. Broad and complex antifungal activity among environmental isolates of lactic acid bacteria. FEMS Microbiol. Lett. 2003, 219, 129–135.
- Ström, K.; Sjögren, J.; Bröberg, A.; Schnürer, J. Lactobacillus plantarum MiLAB 393 produces the antifungal cyclic dipeptides cyclo(L-Phe-L-Pro) and cyclo(L-Phe-trans-4-OH-L-Pro) and phenyllactic acid. Appl. Environ. Microbiol. 2002, 68, 4322–4327.
- Axel, C.; Zannini, E.; Arendt, E.K.; Waters, D.M.; Czerny, M. Quantification of cyclic dipeptides from cultures of Lactobacillus brevis R2D by HRGC/MS using stable isotope dilution assay. Anal. Bioanal. Chem. 2014, 406, 2433–2444.
- Axel, C.; Zannini, E.; Arendt, E.K. Mold spoilage of bread and its biopreservation: A review of current strategies for bread shelf life extension. Crit. Rev. Food Sci. 2017, 57, 3528–3542.
- Kaur, R.; Tiwari, S.K. Isolation, identification and characterization of Pediococcus pentosaceus LB44 and Weissella confusa LM85 for the presence of bacteriocin-like inhibitory substances (BLIS). Microbiology 2016, 85, 540–547.
- Yanagida, F.; Chen, Y.S.; Shinohara, T. Searching for bacteriocin-producing lactic acid bacteria in soil. J. Gen. Appl. Microbiol. 2006, 52, 21–28.
More
Information
Subjects:
Soil Science
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
692
Revisions:
2 times
(View History)
Update Date:
04 Mar 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




