Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Georgio Nemer | -- | 4486 | 2024-02-27 10:56:36 | | | |
| 2 | Lindsay Dong | Meta information modification | 4486 | 2024-02-28 04:28:17 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Nemer, G.; Koubaa, M.; El Chamy, L.; Maroun, R.G.; Louka, N. Colorimetric Whole-Cell Biosensors. Encyclopedia. Available online: https://encyclopedia.pub/entry/55529 (accessed on 07 February 2026).
Nemer G, Koubaa M, El Chamy L, Maroun RG, Louka N. Colorimetric Whole-Cell Biosensors. Encyclopedia. Available at: https://encyclopedia.pub/entry/55529. Accessed February 07, 2026.
Nemer, Georgio, Mohamed Koubaa, Laure El Chamy, Richard G. Maroun, Nicolas Louka. "Colorimetric Whole-Cell Biosensors" Encyclopedia, https://encyclopedia.pub/entry/55529 (accessed February 07, 2026).
Nemer, G., Koubaa, M., El Chamy, L., Maroun, R.G., & Louka, N. (2024, February 27). Colorimetric Whole-Cell Biosensors. In Encyclopedia. https://encyclopedia.pub/entry/55529
Nemer, Georgio, et al. "Colorimetric Whole-Cell Biosensors." Encyclopedia. Web. 27 February, 2024.
Copy Citation
Colorimetric whole-cell biosensors are natural or genetically engineered microorganisms utilized to detect target molecules and ions as indicators of pollutants and biological activity in the environment. Upon detection, within specific concentration ranges which vary depending on the microorganism and its genetic circuitry among other factors, these sensors produce pigments which can be detected with the human eye past certain thresholds and quantified using simple analytical techniques, namely spectrophotometry. These sensors, which can be rendered portable through lyophilization and other methods, provide valuable and reliable substitutes of more demanding analytical ex situ techniques.
whole-cell biosensors
pigments
quantification
qualification
spectrophotometry
1. Introduction
A biosensor is a measurement or quantification system relying on a biological component to recognize target analytes [1]. These devices quantify biological activity or chemical composition through the production of a dose-dependent signal [2]. Whole-cell biosensors (WCBs) are genetically engineered microorganisms capable of detecting and reporting a particular compound or analyte through the emission of a discernable signal using a stimulus-specific reporter system. WCBs are efficient and cost-effective means for obtaining in situ qualitative as well as quantitative information about the medium in which they are introduced. The precision of the readout is governed by a number of factors including the microbial chassis used as a sensor, its resistance to the concentration of the substance being quantified, and the metabolic burden incurred by the synthesis of the output molecule among others. While some biosensors are capable of producing a single readout in response to a specific analyte, others have been engineered to produce distinct concentration-specific outputs [3]. Broadly speaking, these systems utilize a sensing module which detects a specific target (e.g., ion, molecule, or metabolite) and transmits this stimulus to a reporting module which outputs a visible signal. A panoply of characterized biological sensing systems and signaling pathways could be implemented in WCB sensing mechanism design [4]. Transcriptional regulator systems integrate promoters responding to specific environmental constituents linked to engineered gene circuits [5], resulting in the expression or repression of the reporter genes when the promoter-specific compound or protein-ion complex is detected in the medium. Another system relies on a riboswitch comprising an RNA aptamer. Through a conformational change induced by specific metabolite or ligand-binding, the riboswitch may regulate the expression of reporter genes through different mechanisms: halting reporter transcription through the inhibition of antiterminator or the cleavage of mRNA, or activating or repressing translation via the sequestration of the ribosomal binding site (RBS) [6][7]. In essence, WCBs exploit the sensitivity of natural regulatory systems crucial to the survival of microorganisms [8]. They can be utilized for multifarious purposes, such as monitoring natural environments like soil or bodies of water [9][10], screening for high-output strains in biosynthetic industrial settings [11], or providing health data by revealing the amounts of specific micronutrients in human serum among other uses [12].
2. Response of WCBs to Synthetic Molecules
Polychlorinated biphenyls (PCBs) designate a large category of synthetic organic molecules with high hydrophobicity. The potential health complications engendered by PCBs are multitudinous and include neurological conditions, endocrine disruptions, and cancer [13]. Before their worldwide ban through the Stockholm Convention on Persistent Organic Pollutants in 2001, which superseded their 1976 ban in the USA, PCBs were used in a broad range of applications including textiles, construction, transformer oils, and hydraulic equipment. Despite the interruption of their production, the noxious effects of PCBs persist given their capacity to cause complications at remarkably low concentrations. To that end, PCB sensors with sensitivities in the ppb domain had been devised [14][15].
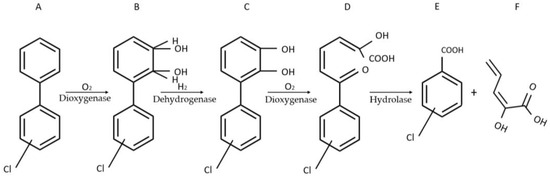
The sensor developed by Gavlasova et al. harnesses the metabolic abilities of Pseudomonas sp. P2 [16]. This strain isolated from a PCB contaminated soil, can, in the presence of biphenyl, convert PCBs into chlorobenzoic acid following a four-step catabolic pathway (Figure 1) [17][18].

Figure 1. Four-step catabolic pathway of PCBs in Pseudomonas sp. P2. Adapted from Gavlasova et al. [16]. (A) biphenyl; (B) 2,3-dihydro-2,3-dihydroxybiphenyl; (C) 2,3-dihydroxybiphenyl; (D) 2-hydroxy-6-oxo-6-phenyl-2,4-hexadienoic acid (HOPDA); (E) benzoic acid; (F) 2-hydroxy-2,4-pentadienoic acid.
In its penultimate step, the pathway engenders the formation of readily observable yellow HOPDA. Through its maximum absorbance at λ = 398 nm, HOPDA produced by Pseudomonas sp. P2 can qualitatively signal the presence of PCBs and semi-quantitatively determine their amounts. While a protocol leveraging HOPDA synthesis had been previously devised by Kuncova et al. [19], suboptimal immobilization of the bacterial cells on glass beads resulted in inaccurate quantification. The innovative aspect of the protocol developed by Gavlasova and colleagues lies in its use of tetramethylorthosilicate (TMOS) to immobilize the homogeneously dispersed WCB on 3 cm petri dishes.
With its myriad chemical compositions and physical properties, soil nurtures numerous interdependent systems of flora and fauna influenced by its attributes and which, in turn, influence its composition. As such, deleterious alterations made to any component of this system can inevitably yield commensurate ripple effects with enduring consequences. Human agricultural activity has entailed the use of a number of pesticides with considerable impact on animals and human health [20]. A number of optical biosensors have been devised to detect organophosphate pesticides through their hydrolysis products, chief among which is 4-nitrophenol [21].
To detect the hydrolysis products of organophosphate pesticides, Chong and Ching successfully produced an E. coli colorimetric WCB using a modified DmpR transcriptional activator, which allowed greater effector specificity and thus higher expression level of its cognate promoter driving the expression of the monomeric red fluorescent protein 1 (mRFP) reporter gene [22]. While lycopene would be an obvious choice should a red pigment be considered as the output signal, the researchers stipulate that the synthesis of lycopene and carotenoids in general is highly influenced by metabolic fluxes. These variabilities might result in inconsistent results from a color intensity perspective and an incubation time standpoint. The use of readily visible red fluorescent proteins, expressed through mRFP1, was deemed likely to engender more dependable results [22]. Transcription regulator DmpR, widely investigated in phenol detection contexts [23][24][25], was used as a sensing module in a wholly mutagenized form. To select 4-nitrophenol sensing mutants most conducive to discernable red fluorescent protein (RFP) synthesis, the researchers used DmpR as a sensing module and mRFP1 as a reporter module. DmpR was mutagenized randomly before reconstituting the sensor plasmid and transforming it into E. coli MG1655. The transformed E. coli strains were cultured on LB agar plates and, after accounting for possible false positives, two 4-nitrophenol-effected DmpR mutants DM01 and DM12 with adequate pigment basal expression were retained. Each of the transcription regulators was cloned onto a plasmid bearing oph, a gene encoding organophosphorus hydrolase or OPH, and mRFP1 to create novel sensor plasmids. Organophosphorus hydrolase enables the hydrolysis of parathion into 4-nitrophenol and thus its detection through DM01 and DM12, followed by the induced synthesis of RFPs. The novel biosensors were able to signal the presence of both hydrolyzed and unhydrolyzed organophosphorus pesticides. The synthesis of organophosphorus hydrolase (OPH) commensurably affected the effectiveness of the sensor and, to overcome this bottleneck, a strong constitutive promoter pTet was selected to drive the expression of the oph gene in the engineered DM01 and DM12 mutant E. coli strains.
3. Detection of Metals by WCBs
3.1. Response of WCBs to Copper
Copper (Cu), among other metal ions such as zinc and manganese, plays a significant role as an enzyme cofactor involved in the catalysis of metabolic activity and the maintenance of cell integrity through osmotic pressure regulation [26]. Despite its considerable utility, copper becomes harmful to humans, animals, and plants alike past respective thresholds, and colorimetric WCBs can provide reasonable data regarding its concentration in water bodies. Several copper metalloregulator systems have been characterized in a number of microorganisms. In Saccharomyces cerevisiae for example, metallothionein encoded in CUP1 enables the chelation of Cu(II) ions and protects the cell from copper poisoning. The copper-dependent DNA-binding protein ACE1 induces CUP1 transcription through the binding of ACE1-Cu(II) complex onto the upstream activation sequence of CUP1 [27].
A colorimetric WCB consisted of an engineered strain of S. cerevisiae with a deleted ADE2 gene, encoding phosphoribosylaminoimidazole carboxylase, and a CUP1 promoter PCUP1 driving the expression of genes at the ADE5,7 locus encoding glycinamide ribotide synthetase and aminoimidazole ribotide synthetase [28][29]. In this strain dubbed BY-ade2-PCUP-ADE5,7, the CUP1 promoter, being inducible mainly by Cu(II) ions [30], leverages the adenine monophosphate pathway altered by ADE2 deletion to enable intracellular accumulation of red pigments in the yeast in high Cu(II) and high O2 environments, resulting in visible color changes commensurate with Cu(II) concentration. The modified S. cerevisiae cells were immobilized in alginate beads and the accumulation of red pigment was found to accurately correlate with Cu(II) concentrations within the 1–100 µM range.
A Cupriavidus metallidurans CH34-based biosensor was developed to quantify Cu(II) ions in aquatic environments through the expression of yellow betaxanthin pigments [31]. C. metallidurans possesses the copSR regulatory system, which grants the microorganism the ability to thrive in environments with high copper levels. To produce an effective biosensor, different promoters of the cop cluster identified in the organism’s genome—PcopT, PcopQ, PcopH, PcopA, and PcopM—were first evaluated in red fluorescent protein biosynthesis assays. To identify the ideal promoter, plasmid backbones bearing copS-copR sequences under the control of native promotor and one of the candidate cop promoters driving the expression of reporter gene rfp were each transformed into C. metallidurans.
3.2. Response of WCBs to Cadmium
Colorimetric WCBs can, in low-resource areas, supplant more complicated and voluminous equipment and enable in situ analyses. Cadmium (Cd) constitutes a source of considerable disruptions within ecosystems given its bioaccumulation, toxicity, and persistence in the environment [32]. A host of WCBs have been devised to detect Cd among other heavy metals and include fluorescent, chemiluminescent, and bioluminescent reporters. A number of Cd-specific metalloregulators have been used in various biosensors to reveal the presence of Cd. Most saliently in the context of WCBs, CadR, which is categorized under the MerR subfamily of metal-ion-sensing transcriptional regulators with variable specificities and was characterized in Pseudomonas aeruginosa [33], regulates its own transcription as well as that of a Cd efflux P-type ATPase CadA, thus making microbial species resistant to high concentrations of the metal.
The Phytoene dehydrogenase (CrtI)-enabled synthesis of red carotenoid pigment deinoxanthin from colorless substrate in Deinococcus radiodurans was leveraged to produce a Cd-selective biosensor [10]. In this instance, D. radiodurans was engineered to exclusively produce deinoxanthin in the presence of Cd(II). To achieve this, the researchers utilized a colorless strain of D. radiodurans dubbed KDH018, whose crtI gene had been previously deleted, as the WCB chassis. To use red deinoxanthin as a reporter, and using E. coli for all genetic manipulations, crtI was cloned from chromosomal DNA of wild-type D. radiodurans onto a pRADZ3 E. coli to D. radiodurans shuttle vector, thus generating pRADI [34].
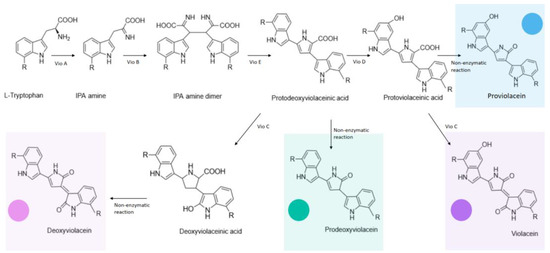
While microorganisms with inherent Cd(II) resistance are conspicuous candidates to explore in this context, model organisms such as E. coli can also be utilized to this end. To detect Cd(II) ions in environmental water with relatively high specificity, a violacein-producing colorimetric biosensor was devised using E. coli as a chassis [35]. The sensory module consisted of the metalloregulator cadR gene, originally characterized in Pseudomonas putida [36], and its divergent cad promoter, whereas the reporter module consisted of a synthetic vioABCDE gene cluster, characterized in Chromobacterium violaceum [37], enabling violacein synthesis (Figure 2) [38].

Figure 2. Enzymatic and non-enzymatic reactions implicated in the violacein biosynthetic pathway encoded by the vioABCDE cluster, as well as the different colored products they yield.
3.3. Response of WCBs to Lead
Environmental lead (Pb) is either naturally occurring or resulting from anthropogenic activity given its use in automotive batteries, telecommunications, construction, and its erstwhile use in pipes and paints among others. The toxicity threshold of lead to humans and animals is quite low, with chronic exposure resulting in anemia, neurotoxicity, and severe renal damage [39]. PbrR, a transcriptional regulator and part of the MerR subfamily, mediates the Pb(II)-induced transcription from its divergent promoter and regulates the pbr operon as part of some microorganisms’ lead detoxification systems [40]. The pbr operon was first identified in Cupriavidus metallidurans CH34 and encodes a particularly comprehensive Pb resistance mechanism which entails transport, efflux, sequestration, precipitation, and biomineralization [41]. As such, components from the pbr operon can and have been successfully utilized in the conception of Pb-sensing WCBs producing various readouts [42][43][44].
The indigoidine reporter module described earlier was also utilized in conjunction with a Pb(II) sensory module comprised of the PbrR transcription regulator and its divergent promoter [45]. The DNA fragments bearing pbrR the pbrR-divergent pbr promoter, borne on the pT-Ppbr plasmid constructed in an earlier study [46], were amplified and cloned into the pT7lac-ind plasmid to yield pPpbr-ind, which was then transformed into E. coli TOP10 competent cells. The output signal is reliably produced 3 h post-incubation with the inductor present, and it can be spectrophotometrically assessed at λ = 600 nm to derive the concentration of bioavailable Pb(II). The authors noted that WCBs harvested at the lag phase were more apt at detecting Pb within the 0.13–4.17 µM range, whereas those harvested at the exponential phase produced the best results within the 0.26–8.3 µM range. While a color change detectable by the human eye can only be observed at 4.17 µM, the method is still conducive to a rapid and reliable spectrophotometric assessment of Pb content in waters suspected of contamination.
Pb(II)-dependent metalloregulator protein PbrR was also used as a component of a sensory module in an E. coli TOP10-based WCB leveraging the violacein biosynthetic pathway [38]. The violacein synthetic pathway was first cloned onto a pET21a plasmid to yield pET-vio. Subsequently, the DNA fragment containing gene pbrR and its divergent promoter Ppbr, borne on the pT-Ppbr plasmid constructed in an earlier study [46], was PCR-amplified and cloned onto the pET-vio plasmid to yield pPpbr-vio used to transform E. coli TOP10. Three hours post-incubation in Pb(II)-containing media, the produced violacein could be extracted using butanol and quantified at λ = 490 nm. A linear relationship between Pb(II) concentration and violacein concentration was observed within the 0.1875–1.5 mM range. Beyond the upper limit of the range, the sensor exhibited qualitative properties up to 24 µM, which was the highest tested concentration. An advantage proffered by the use of violacein in this sensor was the ability to visually detect amounts of Pb(II) in the medium at levels below 0.1875 mM.
Building upon this previously mentioned work, the researchers resorted to subcloning techniques to generate plasmids pPpbr-vioABE, pPpbr-vioABDE, and pPpbr-vioABCE from pPpbr-vio [47]. Each of the plasmids including pPpbr-vio was transformed into E. coli TOP10 to create four WCBs relying on different reporter pigments. The violacein- and deoxyviolacein-producing strains were retained for later assays given that the expected pigment output was produced as opposed to what was observed with the remaining strains. This occurrence can be attributed to the instability of prodeoxyviolacein and proviolacein intermediates as well as the highly branched metabolic pathway culminating in violacein synthesis [48]. The deoxyviolacein-based biosensor displayed considerable efficacy and a narrow dose-dependent response across a broad range of Pb(II) concentrations spanning 2.93–6000 nM, whereas the violacein-based one exhibited a less narrow dose-dependent response between 2.93 nM and 750 nM.
3.4. Response of WCBs to Mercury
The highly toxic nature of bioaccumulative mercury (Hg) coupled with its relative abundance in the environment make it a veritable hazard to public health. Several bacteria have developed resistance mechanisms to the metal. These detoxification mechanisms, enabled by an inducible set of genes arranged in a single MerR operon under the control of the metal-sensing MerR protein [49], have been exploited in a number of contexts to create crucial biosensors enabling the safeguarding of human and animal health.
The polyvalence of the indigoidine reporter module is further evinced in its robustness at signaling the presence of Hg(II) in an E. coli-based WCB [45]. This was achieved by coupling the reporter module with a sensor module consisting of the MerR gene encoding the metalloregulator protein MerR and its divergent mer promoter region, which had been synthetically produced and introduced into a pET-21a plasmid to generate Ppmer [50]. The DNA fragments contained in Ppmer were PCR amplified and cloned into the previously referenced pT7lac-ind plasmid to generate pPmer-ind, which was then transformed into E. coli TOP10. WCBs harvested at the exponential phase of bacterial development were found to reliably signal and quantify Hg(II), which induced a dose-responsive indigoidine biosynthesis at a concentration range spanning 0.008–0.52 µM, with the color of the pigment exceeding the human eye detection threshold at Hg(II) concentrations above 0.033 µM.
The violacein reporter developed by Hui et al. and utilized to detect Pb in an earlier work [38] was repurposed in the creation of a Hg(II) biosensor [51]. The Ppmer sensor module which was constructed by Zhang et al. [50] was also utilized in this undertaking. To assemble the sensory and reporter apparatuses, the researchers PCR amplified the DNA fragment containing merR gene and its divergent mer promoter from the Ppmer plasmid and cloned them into pET-vio to generate pPmer-vio. The recombinant plasmid was transformed into E. coli TOP10 to yield a Hg-sensing WCB which would respond to Hg by producing the violet pigment violacein. A dependable readout from WCB cells harvested during the exponential phase was obtained 5 h post-induction, and it exhibited a dose-dependent pigment-based response to Hg(II) in the range of 0.78–12.5 µM. Beyond this upper limit, violacein synthesis reportedly decreased as a result of toxicity. Inductive amounts of Hg(II) equal to and beyond 6.25 µM incurred the production of enough pigment to be detected by the human eye post-extraction using butanol. Sensors harvested at the lag phase exhibited a dose-dependent response to Hg(II) within the 0–0.12 µM range, thus allowing for the quantification of more minute amounts of Hg(II). Violacein was visible with the human eye post-extraction at Hg(II) concentrations withing the 0.006–0.098 µM range and the intensity of the violet color diminished past the Hg(II) concentration of 0.024 µM due to cytotoxic effects.
Transcription regulator MerR has also been used as a sensor module in a P. aeruginosa WCB employing reporter genes phzM and phzS, encoding for the enzymes methyltransferase and flavin-containing monooxygenase, respectively [52]. These enzymes catalyze the synthesis of pyomelanin [53], a red–brown pigment with potent antioxidative properties protecting microorganisms from oxidative stress [54]. A recombinant plasmid carrying merR under the control of native promoter and the phzM and phzS genes under the control of the mer promoter, was transformed into P. aeruginosa PAO1. The WCB worked well within a broad pH range, proved to be highly selective by responding poorly to other metal ions and produced a dose-dependent response to Hg(II) between 25 and 1000 nM. Prior to spectrophotometric quantification of pyocyanin to derive Hg(II) concentrations, the hydrophobic pigment must be extracted from the cells using chloroform and hydrochloric acid.
3.5. Response of WCBs to Arsenic
Over one hundred million people are effected by arsenic (As) water contamination across the world and are thus prone to developing skin lesions and gastrointestinal distress in case of chronic exposure to low doses, although high concentrations pose a much greater toxicity risk to human health [55]. A number of WCBs were elaborated utilizing the ars operon, which consists of two regulatory genes (arsR and arsD) and three structural genes (arsA, arsB, and arsC) and contributes to arsenite and arsenate resistance by detoxifying the cell [56]. Highly contaminated areas include Indian and Bangladeshi industrial zones, and remediation must be enabled by access to cheap and dependable technologies.
The bright colors of carotenoid pigments spheroiden and spheroidenone were exploited in the creation of an arsenite biosensor [57]. In its wild form, photosynthetic bacterium Rhodovulum sulfidophilum produces the red pigment spheroidenone in semi-aerobic conditions via the spheroidone pathway enabled by genes crtF and crtA. The former encodes for O-methyltransferase which acts upon the C-1 hydroxy group of demethylspheroidene resulting in the synthesis of yellow spheroiden, and the latter codes for a monooxygenase subjecting the spheroiden produced in the earlier step to a C-2 ketolation thus yielding red spheroidenone. As such, a mutant strain with the crtA gene deleted, such as R. sulfidophilum CDM2, would accumulate yellow pigments. To create the arsenite sensor, Fujimoto et al. relied on a strategy predicated on a color shift from yellow to red using R. sulfidophilum CDM2. To that end, they constructed a reporter module consisting of a promoter-less fragment of the crtA gene which was cloned onto a broad-host-range plasmid pRK415 together with the E. coli-derived sensory module comprising the arsenite responsive E. coli DNA fragment containing the operator/promoter of the ars operon (O/pars) as well as the the arsR repressor. The recombinant plasmid, pSENSE-As, was transformed into E. coli S17-1 and transferred into R. sulfidophilum CDM2 through conjugation. Preliminary assessments confirmed that E. coli O/Pars was recognizable by CDM2 RNA polymerase and that no transcription repression by an endogenous protein occurred in CDM2.
4. Biomonitoring and Control
4.1. High-Level Producer Detection
Lysine is an amino acid with considerable importance in the context of human and animal nutrition and is the second most abundantly produced essential nutrient worldwide [58][59]. Corynebacterium glutamicum is an effective production platform of L-lysine and other amino acids, and engineered strains have been turned into industrial workhorses specially created for this purpose [60].
A notable drawback on pSenLys-based sensors is their inability to accurately report the overproduction of a specific amino acid among lysine, histidine, and arginine. The non-specificity of pSenLys prompted the exploration of different avenues. Liu et al. detailed the development of a C. glutamicum based colorimetric WCB with greater lysine specificity, utilizing lycopene as a reporter pigment [11]. In response to L-Lys, LysG activates the expression of crtI encoding phytoene desaturase which then catalyzes the production of lycopene with the characteristic red color. To that end, a C. glutamicum mutant strain, deficient of the carotenogenic gene cluster crtIYe/fEb, crtB2I21/2, and LysEG was first generated as a sensor chassis. To construct the plasmid, transcriptional regulator lysG; its binding site region lysE promoter; and the phytoene desaturase gene crtI were amplified using the genome of C. glutamicum as a template. The expression cassette was fused using overlap extension PCR and cloned into a pTRCmob vector plasmid dubbed pSensorI. The sensor plasmid was transformed into C. glutamicum WT-ΔlysEGΔcrtIYEbB22 and this transformed strain bearing pSensorI served as a control as other optimizations were implemented. To remedy the poor specificity of lysG and diminish false positives induced by docking of L-histidine and L-arginine, LysG was subjected to site-directed saturation mutagenesis to screen for mutants with reduced affinity to L-histidine and L-arginine. Substitutions at positions 123 and 125—where L-glutamate was substituted with L-L-tyrosine and where L-glutamate was replaced with L-alanine, respectively—were found to confer the modified binding site lysG* a drastically reduced affinity to L-arginine and L-histidine and an uncompromised colorimetric linear response to L-lysine. To increase the range of dose-responsiveness up from the reported 40 mM limit, the researchers resorted to promoter engineering of pLysE and 5 promoters were screened. Promoter pLysE-3 was selected as the most apt candidate and was found to engender an increased range of responsiveness of up to 300 mM. To enhance the performance of the sensor from a color-rendering standpoint and thus facilitate overproducer detection, the CrtR transcriptional regulator, which is known to repress the crt operon [61], was deleted by electroporating a suicide plasmid into WT-ΔlysEGΔcrtIYEbB2 to construct WT-ΔlysEGΔcrtIYEbB2R.
4.2. Pathogen Detection
N-acyl homoserine lactone (AHL) is a signal molecule utilized by a number of gram-negative bacteria for cell-to-cell communication as part of the quorum sensing mechanism. The utility of WCBs leveraging the quorum sensing apparatus of microorganisms has a number of benefits namely signaling the presence of possibly pathogenic species [62], monitoring bacterial populations in bioreactor settings [63], and modulating the microbial composition of a medium [64].
N-butyryl-L-homoserine lactone (BHL) is an AHL and a small diffusible signaling molecule implicated in quorum sensing, the control of gene expression, and cellular metabolism [65]. To detect minute amounts of BHL within a wide concentration ambit, Yong and Zhong developed a P. aeruginosa-based biosensor [66]. The researchers used strain P. aeruginosa CGMCC 1.860, which is naturally capable of producing blue–green pigments upon detecting BHL. This is achieved through the RhlR-RhlI quorum-sensing system, which comprises the transcription activator protein RhlR and the BHL synthase RhlI [67]. To create a biosensor, the researchers deleted the rhlIR gene cluster, thus creating P. aeruginosa ΔrhlIR, and overexpressed rhlR through multi-copy plasmids. As such, the bacteria regained the capacity of sensing BHL while avoiding the production of the analyte by endogenous activity. The recombinant biosensor strain is thus capable of producing the pigment upon sensing of exogenous BHL which can diffuse in the cell and be recognized by the RhlR regulator. Upon BHL binding, this transcription regulator activates the expression of pigment synthases. The resulting WCB whose pigment output can be extracted using chloroform and quantified at λ = 299 nm exhibited dose-dependent pigment production within the 0.11–49.7 µM AHL range.
4.3. Micronutrient Quantification
Micronutrient deficiencies are significant concern of global ambit although gauging the veritable magnitude of the issue remains challenging [68]. In remote settings and in impoverished parts of the world, access to reliable testing is limited due to elevated costs and logistical difficulties. Colorimetric WCBs as part of field-ready kits can be handled by agents with minimal training to provide in situ testing in remote areas, identify micronutrient deficiencies, and help remedy health complications quickly and reliably.
Zinc is an essential micronutrient; deficiencies have incurred public health burdens of significant magnitude, and one billion people across the world are presumed to be at risk of zinc deficiency [69]. Efforts to provide access to impoverished and remote areas of the world have yielded the development of colorimetric biosensors.
In the context of early efforts to develop a colorimetric biosensor compatible with zinc serum levels, Watstein and Styczynski generated an E. coli-DH10B-based sensor capable of producing three different reporters: violacein, lycopene, and β-carotene [12]. A violacein operon was cloned onto a plasmid bearing zinc-responsive transcription regulator ZntR, proprietary ribosomal binding sites, and the gene encoding the Zur metalloregulator protein, which acts as a zinc-responsive repressor. Gene cluster vioABCDE was placed under the control of a PznuC repressor actuated by Zur–zinc complexes.
5. Conclusions
The development of whole-cell biosensors keeps up with the pace of broad ranging advancements in genetic engineering and practically puts to use novel approaches stemming from recent advancements. As such, it allows for more nuance to materialize and for a greater understanding of processes and mechanisms to be gleaned. WCBs are a highly versatile platform enabling the development of accessible and inexpensive analytical devices which, in some circumstances, replace their less portable laboratory analogues [70]. They can be adapted to a considerable range of analytes that grows as more regulatory as well as biosynthetic pathways are characterized. Moreover, they benefit from a wide selection of thoroughly understood microbial chassis to transform based on the biosensor’s purpose. Indeed, the caveat of these assessments being conducted in highly controlled settings must be borne in mind. In effect, they were mostly undertaken using solutions of known analyte concentrations and compositions. Their effectiveness in the field may be less pronounced due to a host of causes including the general complexity of natural matrices which may contain compounds of bacterial origin or otherwise which may affect their performance or result in false positives and negatives.
References
- Bousse, L. Whole Cell Biosensors. Sens. Actuators B Chem. 1996, 34, 270–275.
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to Biosensors. Essays Biochem. 2016, 60, 1–8.
- Wang, B.; Barahona, M.; Buck, M. A Modular Cell-Based Biosensor Using Engineered Genetic Logic Circuits to Detect and Integrate Multiple Environmental Signals. Biosens. Bioelectron. 2013, 40, 368–376.
- Wen, K.Y.; Rutter, J.W.; Barnes, C.P.; Dekker, L. Fundamental Building Blocks of Whole-Cell Biosensor Design. Handb. Cell Biosens. 2020, 1–23.
- Khalil, A.S.; Collins, J.J. Synthetic Biology: Applications Come of Age. Nat. Rev. Genet. 2010, 11, 367–379.
- Hossain, G.S.; Saini, M.; Miyake, R.; Ling, H.; Chang, M.W. Genetic Biosensor Design for Natural Product Biosynthesis in Microorganisms. Trends Biotechnol. 2020, 38, 797–810.
- Serganov, A.; Nudler, E. A Decade of Riboswitches. Cell 2013, 152, 17–24.
- Wang, B.; Barahona, M.; Buck, M. Amplification of Small Molecule-Inducible Gene Expression via Tuning of Intracellular Receptor Densities. Nucleic Acids Res. 2015, 43, 1955–1964.
- Hui, C.Y.; Guo, Y.; Gao, C.X.; Li, H.; Lin, Y.R.; Yun, J.P.; Chen, Y.T.; Yi, J. A Tailored Indigoidine-Based Whole-Cell Biosensor for Detecting Toxic Cadmium in Environmental Water Samples. Environ. Technol. Innov. 2022, 27, 102511.
- Joe, M.H.; Lee, K.H.; Lim, S.Y.; Im, S.H.; Song, H.P.; Lee, I.S.; Kim, D.H. Pigment-Based Whole-Cell Biosensor System for Cadmium Detection Using Genetically Engineered Deinococcus Radiodurans. Bioprocess Biosyst. Eng. 2012, 35, 265–272.
- Liu, J.; Xu, J.Z.; Rao, Z.M.; Zhang, W.G. An Enzymatic Colorimetric Whole-Cell Biosensor for High-Throughput Identification of Lysine Overproducers. Biosens. Bioelectron. 2022, 216, 114681.
- Watstein, D.M.; Styczynski, M.P. Development of a Pigment-Based Whole-Cell Zinc Biosensor for Human Serum. ACS Synth. Biol. 2018, 7, 267–275.
- Carpenter, D.O. Polychlorinated Biphenyls (PCBs): Routes of Exposure and Effects on Human Health. Rev. Environ. Health 2006, 21, 1–23.
- Přibyl, J.; Hepel, M.; Skládal, P. Piezoelectric Immunosensors for Polychlorinated Biphenyls Operating in Aqueous and Organic Phases. Sens. Actuators B Chem. 2006, 113, 900–910.
- Shimomura, M.; Nomura, Y.; Zhang, W.; Sakino, M.; Lee, K.H.; Ikebukuro, K.; Karube, I. Simple and Rapid Detection Method Using Surface Plasmon Resonance for Dioxins, Polychlorinated Biphenylx and Atrazine. Anal. Chim. Acta 2001, 434, 223–230.
- Gavlasova, P.; Kuncova, G.; Kochankova, L.; Mackova, M. Whole Cell Biosensor for Polychlorinated Biphenyl Analysis Based on Optical Detection. Int. Biodeterior. Biodegrad. 2008, 62, 304–312.
- Dercová, K.; Baláž, Š.; Vrana, B.; Tandlich, R. Aerobic Biodegradation of Polychlorinated Biphenyls (PCBs). In The Utilization of Bioremediation to Reduce Soil Contamination: Problems and Solutions; Springer: Dordrecht, The Netherlands, 2003; pp. 95–113.
- Nováková, H.; Vošahlíková, M.; Pazlarová, J.; Macková, M.; Burkhard, J.; Demnerová, K. PCB Metabolism by Pseudomonas Sp. P2. Int. Biodeterior. Biodegrad. 2002, 50, 47–54.
- Kuncova, G.; Berkova, D.; Burkhard, J.; Demnerova, K.; Pazlarova, J.; Triska, J.; Vrchotova, N. Optical Detection of Polychiorinated Biphenyls. In Environmental Monitoring and Remediation Technologies II; Vo-Dinh, T., Spellicy, R.L., Eds.; SPIE: Bellingham, WA, USA, 1999; Volume 3853, pp. 72–80.
- Mulchandani, A.; Rajesh. Microbial Biosensors for Organophosphate Pesticides. Appl. Biochem. Biotechnol. 2011, 165, 687–699.
- Kumar, J.; Jha, S.K.; D’Souza, S.F. Optical Microbial Biosensor for Detection of Methyl Parathion Pesticide Using Flavobacterium Sp. Whole Cells Adsorbed on Glass Fiber Filters as Disposable Biocomponent. Biosens. Bioelectron. 2006, 21, 2100–2105.
- Chong, H.; Ching, C.B. Development of Colorimetric-Based Whole-Cell Biosensor for Organophosphorus Compounds by Engineering Transcription Regulator DmpR. ACS Synth. Biol. 2016, 5, 1290–1298.
- Wise, A.A.; Kuske, C.R. Generation of Novel Bacterial Regulatory Proteins That Detect Priority Pollutant Phenols. Appl. Environ. Microbiol. 2000, 66, 163–169.
- Pavel, H.; Forsman, M.; Shingler, V. An Aromatic Effector Specificity Mutant of the Transcriptional Regulator DmpR Overcomes the Growth Constraints of Pseudomonas Sp. Strain CF600 on Para-Substituted Methylphenols. J. Bacteriol. 1994, 176, 7550–7557.
- Campos, V.L.; Zaror, C.A.; Mondaca, M.A. Detection of Chlorinated Phenols in Kraft Pulp Bleaching Effluents Using DmpR Mutant Strains. Bull. Environ. Contam. Toxicol. 2004, 73, 666–673.
- Nies, D.H. Heavy Metal-Resistant Bacteria as Extremophiles: Molecular Physiology and Biotechnological Use of Ralstonia Sp. CH34. Extremophiles 2000, 4, 77–82.
- Knight, S.A.B.; Tamai, K.T.; Kosman, D.J.; Thiele, D.J. Identification and Analysis of a Saccharomyces Cerevisiae Copper Homeostasis Gene Encoding a Homeodomain Protein. Mol. Cell. Biol. 1994, 14, 7792–7804.
- Vopálenská, I.; Váchová, L.; Palková, Z. New Biosensor for Detection of Copper Ions in Water Based on Immobilized Genetically Modified Yeast Cells. Biosens. Bioelectron. 2015, 72, 160–167.
- Henikoff, S. The Saccharomyces Cerevisiae ADE5,7 Protein Is Homologous to Overlapping Drosophila Melanogaster Gart Polypeptides. J. Mol. Biol. 1986, 190, 519–528.
- Shetty, R.S.; Deo, S.K.; Liu, Y.; Daunert, S. Fluorescence-Based Sensing System for Copper Using Genetically Engineered Living Yeast Cells. Biotechnol. Bioeng. 2004, 88, 664–670.
- Chen, P.H.; Lin, C.; Guo, K.H.; Yeh, Y.C. Development of a Pigment-Based Whole-Cell Biosensor for the Analysis of Environmental Copper. RSC Adv. 2017, 7, 29302–29305.
- Kumar, A.; Subrahmanyam, G.; Mondal, R.; Cabral-Pinto, M.M.S.; Shabnam, A.A.; Jigyasu, D.K.; Malyan, S.K.; Fagodiya, R.K.; Khan, S.A.; Yu, Z.G. Bio-Remediation Approaches for Alleviation of Cadmium Contamination in Natural Resources. Chemosphere 2021, 268, 128855.
- Hui, C.Y.; Hu, S.Y.; Li, L.M.; Yun, J.P.; Zhang, Y.F.; Yi, J.; Zhang, N.X.; Guo, Y. Metabolic Engineering of the Carotenoid Biosynthetic Pathway toward a Specific and Sensitive Inorganic Mercury Biosensor. RSC Adv. 2022, 12, 36142–36148.
- Meima, R.; Lindstrom, M.E. Characterization of the Minimal Replicon of a Cryptic Deinococcus Radiodurans SARK Plasmid and Development of Versatile Escherichia Coli-D. Radiodurans Shuttle Vectors. Appl. Environ. Microbiol. 2000, 66, 3856–3867.
- Hui, C.Y.; Guo, Y.; Li, H.; Gao, C.X.; Yi, J. Detection of Environmental Pollutant Cadmium in Water Using a Visual Bacterial Biosensor. Sci. Rep. 2022, 12, 1–11.
- Lee, S.W.; Glickmann, E.; Cooksey, D.A. Chromosomal Locus for Cadmium Resistance in Pseudomonas Putida Consisting of a Cadmium-Transporting ATPase and a MerR Family Response Regulator. Appl. Environ. Microbiol. 2001, 67, 1437–1444.
- August, P.R.; Grossman, T.H.; Minor, C.; Draper, M.P.; MacNeil, I.A.; Pemberton, J.M.; Call, K.M.; Holt, D.; Osbourne, S. Sequence Analysis and Functional Characterization of the Violacein Biosynthetic Pathway from Chromobacterium Violaceum. J. Mol. Microbiol. Biotechnol. 2000, 2, 513–519.
- Hui, C.Y.; Guo, Y.; Liu, L.; Zhang, N.X.; Gao, C.X.; Yang, X.Q.; Yi, J. Genetic Control of Violacein Biosynthesis to Enable a Pigment-Based Whole-Cell Lead Biosensor. RSC Adv. 2020, 10, 28106–28113.
- Shetty, R.S.; Deo, S.K.; Shah, P.; Sun, Y.; Rosen, B.P.; Daunert, S. Luminescence-Based Whole-Cell-Sensing Systems for Cadmium and Lead Using Genetically Engineered Bacteria. Anal. Bioanal. Chem. 2003, 376, 11–17.
- Borremans, B.; Hobman, J.L.; Provoost, A.; Brown, N.L.; Van Der Lelie, D. Cloning and Functional Analysis of the Pbr Lead Resistance Determinant of Ralstonia Metallidurans CH34. J. Bacteriol. 2001, 183, 5651–5658.
- Hui, C.; Ma, B.; Wang, Y.; Yang, X.; Cai, J. Designed Bacteria Based on Natural Pbr Operons for Detecting and Detoxifying Environmental Lead: A Mini-Review. Ecotoxicol. Environ. Saf. 2023, 267, 115662.
- Bereza-Malcolm, L.; Aracic, S.; Franks, A.E. Development and Application of a Synthetically-Derived Lead Biosensor Construct for Use in Gram-Negative Bacteria. Sensors 2016, 16, 2174.
- Nourmohammadi, E.; Hosseinkhani, S.; Nedaeinia, R.; Khoshdel-Sarkarizi, H.; Nedaeinia, M.; Ranjbar, M.; Ebrahimi, N.; Farjami, Z.; Nourmohammadi, M.; Mahmoudi, A.; et al. Construction of a Sensitive and Specific Lead Biosensor Using a Genetically Engineered Bacterial System with a Luciferase Gene Reporter Controlled by Pbr and CadA Promoters. Biomed. Eng. Online 2020, 19, 1–13.
- Jia, X.; Zhao, T.; Liu, Y.; Bu, R.; Wu, K. Gene Circuit Engineering to Improve the Performance of a Whole-Cell Lead Biosensor. FEMS Microbiol. Lett. 2018, 365, 1–8.
- Hui, C.Y.; Guo, Y.; Li, L.M.; Liu, L.; Chen, Y.T.; Yi, J.; Zhang, N.X. Indigoidine Biosynthesis Triggered by the Heavy Metal-Responsive Transcription Regulator: A Visual Whole-Cell Biosensor. Appl. Microbiol. Biotechnol. 2021, 105, 6087–6102.
- Guo, Y.; Hui, C.Y.; Liu, L.; Zheng, H.Q.; Wu, H.M. Improved Monitoring of Low-Level Transcription in Escherichia Coliby a β-Galactosidase α-Complementation System. Front. Microbiol. 2019, 10, 1–12.
- Hui, C.Y.; Guo, Y.; Zhu, D.L.; Li, L.M.; Yi, J.; Zhang, N.X. Metabolic Engineering of the Violacein Biosynthetic Pathway toward a Low-Cost, Minimal-Equipment Lead Biosensor. Biosens. Bioelectron. 2022, 214, 114531.
- Zhao, E.M.; Suek, N.; Wilson, M.Z.; Dine, E.; Pannucci, N.L.; Gitai, Z.; Avalos, J.L.; Toettcher, J.E. Light-Based Control of Metabolic Flux through Assembly of Synthetic Organelles. Nat. Chem. Biol. 2019, 15, 589–597.
- Helmann, J.D.; Ballard, B.T.; Walsh, C.T. The MerR Metalloregulatory Protein Binds Mercuric Ion as a Tricoordinate, Metal-Bridged Dimer. Science 1990, 247, 946–948.
- Zhang, N.X.; Guo, Y.; Li, H.; Yang, X.Q.; Gao, C.X.; Hui, C.Y. Versatile Artificial Mer Operons in Escherichia Coli towards Whole Cell Biosensing and Adsorption of Mercury. PLoS ONE 2021, 16, 1–14.
- Guo, Y.; Hui, C.Y.; Liu, L.; Chen, M.P.; Huang, H.Y. Development of a Bioavailable Hg(II) Sensing System Based on MerR-Regulated Visual Pigment Biosynthesis. Sci. Rep. 2021, 11, 1–13.
- Huang, L.; Huang, Y.; Lou, Y.; Qian, H.; Xu, D.; Ma, L.; Jiang, C.; Zhang, D. Pyocyanin-Modifying Genes PhzM and PhzS Regulated the Extracellular Electron Transfer in Microbiologically-Influenced Corrosion of X80 Carbon Steel by Pseudomonas Aeruginosa. Corros. Sci. 2020, 164, 108355.
- Wang, D.; Zheng, Y.; Fan, X.; Xu, L.; Pang, T.; Liu, T.; Liang, L.; Huang, S.; Xiao, Q. Visual Detection of Hg2+ by Manipulation of Pyocyanin Biosynthesis through the Hg2+-Dependent Transcriptional Activator MerR in Microbial Cells. J. Biosci. Bioeng. 2020, 129, 223–228.
- Ketelboeter, L.M.; Bardy, S.L. Methods to Inhibit Bacterial Pyomelanin Production and Determine the Corresponding Increase in Sensitivity to Oxidative Stress. J. Vis. Exp. 2015, 2015, 1–9.
- De Mora, K.; Joshi, N.; Balint, B.L.; Ward, F.B.; Elfick, A.; French, C.E. A PH-Based Biosensor for Detection of Arsenic in Drinking Water. Anal. Bioanal. Chem. 2011, 400, 1031–1039.
- Wu, J.; Rosen, B.P. The ArsR Protein Is a Trans-acting Regulatory Protein. Mol. Microbiol. 1991, 5, 1331–1336.
- Fujimoto, H.; Wakabayashi, M.; Yamashiro, H.; Maeda, I.; Isoda, K.; Kondoh, M.; Kawase, M.; Miyasaka, H.; Yagi, K. Whole-Cell Arsenite Biosensor Using Photosynthetic Bacterium Rhodovulum Sulfidophilum: Rhodovulum Sulfidophilum as an Arsenite Biosensor. Appl. Microbiol. Biotechnol. 2006, 73, 332–338.
- Félix, F.K. do C.; Letti, L.A.J.; Vinícius de Melo Pereira, G.; Bonfim, P.G.B.; Soccol, V.T.; Soccol, C.R. L-Lysine Production Improvement: A Review of the State of the Art and Patent Landscape Focusing on Strain Development and Fermentation Technologies. Crit. Rev. Biotechnol. 2019, 39, 1031–1055.
- Cheng, J.; Chen, P.; Song, A.; Wang, D.; Wang, Q. Expanding Lysine Industry: Industrial Biomanufacturing of Lysine and Its Derivatives. J. Ind. Microbiol. Biotechnol. 2018, 45, 719–734.
- Ikeda, M. Lysine Fermentation: History and Genome Breeding. In Advances in Biochemical Engineering/Biotechnology; Springer: Tokyo, Japan, 2016; Volume 123, pp. 73–102.
- Henke, N.A.; Wendisch, V.F. Improved Astaxanthin Production with Corynebacterium Glutamicum by Application of a Membrane Fusion Protein. Mar. Drugs 2019, 17, 621.
- Kumari, A.; Pasini, P.; Daunert, S. Detection of Bacterial Quorum Sensing N-Acyl Homoserine Lactones in Clinical Samples. Anal. Bioanal. Chem. 2008, 391, 1619–1627.
- Yeon, K.M.; Cheong, W.S.; Oh, H.S.; Lee, W.N.; Hwang, B.K.; Lee, C.H.; Beyenal, H.; Lewandowski, Z. Quorum Sensing: A New Biofouling Control Paradigm in a Membrane Bioreactor for Advanced Wastewater Treatment. Environ. Sci. Technol. 2009, 43, 380–385.
- Valle, A.; Bailey, M.J.; Whiteley, A.S.; Manefield, M. N-Acyl-L-Homoserine Lactones (AHLs) Affect Microbial Community Composition and Function in Activated Sludge. Environ. Microbiol. 2004, 6, 424–433.
- Song, S.; Jia, Z.; Xu, J.; Zhang, Z.; Bian, Z. N-Butyryl-Homoserine Lactone, a Bacterial Quorum-Sensing Signaling Molecule, Induces Intracellular Calcium Elevation in Arabidopsis Root Cells. Biochem. Biophys. Res. Commun. 2011, 414, 355–360.
- Yong, Y.C.; Zhong, J.J. A Genetically Engineered Whole-Cell Pigment-Based Bacterial Biosensing System for Quantification of N-Butyryl Homoserine Lactone Quorum Sensing Signal. Biosens. Bioelectron. 2009, 25, 41–47.
- Sun, G.X.; Zhou, W.Q.; Zhong, J.J. Organotin Decomposition by Pyochelin, Secreted by Pseudomonas Aeruginosa Even in an Iron-Sufficient Environment. Appl. Environ. Microbiol. 2006, 72, 6411–6413.
- Stevens, G.A.; Beal, T.; Mbuya, M.N.N.; Luo, H.; Neufeld, L.M.; Addo, O.Y.; Adu-Afarwuah, S.; Alayón, S.; Bhutta, Z.; Brown, K.H.; et al. Micronutrient Deficiencies among Preschool-Aged Children and Women of Reproductive Age Worldwide: A Pooled Analysis of Individual-Level Data from Population-Representative Surveys. Lancet Glob. Health 2022, 10, e1590–e1599.
- Wessells, K.R.; Brown, K.H. Estimating the Global Prevalence of Zinc Deficiency: Results Based on Zinc Availability in National Food Supplies and the Prevalence of Stunting. PLoS ONE 2012, 7.
- Moraskie, M.; Roshid, H.; Connor, G.O.; Dikici, E.; Zingg, J.; Deo, S.; Daunert, S. Microbial Whole-Cell Biosensors: Current Applications, Challenges, and Future Perspectives. Biosens. Bioelectron. 2022, 191, 113359.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
533
Revisions:
2 times
(View History)
Update Date:
28 Feb 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




