Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Gkikas Magiorkinis | -- | 2381 | 2024-02-23 16:40:53 | | | |
| 2 | Wendy Huang | Meta information modification | 2381 | 2024-02-26 05:23:37 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Mantovani, F.; Kitsou, K.; Magiorkinis, G. Endogenous Retroviruses Expression Regulation Mechanisms. Encyclopedia. Available online: https://encyclopedia.pub/entry/55400 (accessed on 10 March 2026).
Mantovani F, Kitsou K, Magiorkinis G. Endogenous Retroviruses Expression Regulation Mechanisms. Encyclopedia. Available at: https://encyclopedia.pub/entry/55400. Accessed March 10, 2026.
Mantovani, Federica, Konstantina Kitsou, Gkikas Magiorkinis. "Endogenous Retroviruses Expression Regulation Mechanisms" Encyclopedia, https://encyclopedia.pub/entry/55400 (accessed March 10, 2026).
Mantovani, F., Kitsou, K., & Magiorkinis, G. (2024, February 23). Endogenous Retroviruses Expression Regulation Mechanisms. In Encyclopedia. https://encyclopedia.pub/entry/55400
Mantovani, Federica, et al. "Endogenous Retroviruses Expression Regulation Mechanisms." Encyclopedia. Web. 23 February, 2024.
Copy Citation
Human endogenous retroviruses (HERVs) are the result of retroviral infections acquired millions of years ago; nowadays, they compose around 8% of human DNA. Multiple mechanisms have been employed for endogenous retroviral deactivation, rendering replication and retrotransposition defective, while some of them have been co-opted to serve host evolutionary advantages. A pleiad of mechanisms retains the delicate balance of HERV expression in modern humans. Thus, epigenetic modifications, such as DNA and histone methylation, acetylation, deamination, chromatin remodeling, and even post-transcriptional control are recruited.
HERVs
expression
regulation
methylation
DNA
histone
acetylation
deamination
chromatin
1. Introduction
Human endogenous retroviruses (HERVs) are the result of retroviral endogenization, which happened millions of years ago and currently comprise about 8% of the human genome. HERVs are classified as retrotransposons; they originally utilized reverse transcription for their duplication and reintegration in other sites of the human genome [1]. In humans, ERVs have lost their capacity for replication and retrotransposition [2], while the majority of their loci have lost the coding ability after the accumulation of point mutations, including non-sense mutations, leading to premature stop codons and frameshifts [3]. Still, though, some LTRs have retained intact ORFs with coding capacity [3]. The full-length provirus resembles the typical structure of a simple retrovirus, with the two LTRs flanking the gag, pro, pol, and env genes (reviewed in Mao et al., 2021 [4]) (Figure 1), coding for matrix, capsid, and nucleocapsid proteins (gag), protease (pro), reverse transcriptase and integrase (pol), and the envelope (env) [4][5]. HERVs have originally been classified in families, the nomenclature of which is defined by the tRNA primer binding site (PBS) at 5′ LTR of gag [6]; however, such categorization leads to problems regarding the actual phylogenetic relations among members of the same family [7].
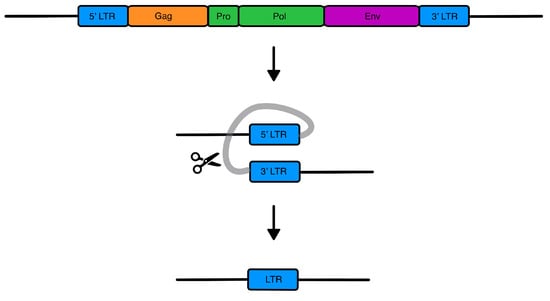
Figure 1. HERV genes and solo-LTR formation. The HERV provirus contains four main genes: Gag, encoding for the matrix, nucleocapsid, and capsid proteins; Pro, encoding for the protease; Pol, encoding the retrotranscriptase and integrase; and Env, encoding for the envelope. Flanking the genes are two LTRs with enhancer and promoter activity. Solo LTRs are created through homologous recombination, while the internal part containing the genes is lost.
Throughout evolution, certain HERV-originating proteins appear to have been co-opted by mammals to acquire new functions to the benefit of the host [8], most importantly the syncytins (HERV-W envelope: syncytin-1, HERV-FRD envelope: syncytin-2) that majorly contribute to the successful placentation [9]. Furthermore, HERV expression, namely HERV-H, HERV-L, and HERV-K [10], is higher in stem cells and during embryo development, contrary to differentiated cells, suggesting a potential role in the maintenance of the pluripotency state [10]. Additionally, full-length HERV sequences and mainly LTRs can also function as regulatory elements with promoter, enhancer, and repressor activity, poly-A signals, and alternative splicing sites in the human genome [11][12][13].
To maintain the physiological functions of the hosts and control HERV expression at levels that suit each cell type and developmental stage, multiple mechanisms have been developed in the course of evolutionary history. In the following sections, the main mechanisms that aid in the maintenance of the hosts’ homeostasis will be explored.
2. Methylation
Methylation, both regarding the retrovirally originating DNA sequences and in the frames of histone modifications, is employed for HERV transcriptional repression, with the former being mainly a characteristic of the somatic tissues, while the histone 3 lysine 9 trimethylation (H3K9me3) is mainly used as a silencing mechanism in the embryonic stem cells (ESCs) [14]. The first mechanism is used to avoid cell reprogramming, while histone methylation is more commonly associated with the prevention of unfavorable gene activation [15]. Strikingly, in the differentiating tissues, lower methylation levels and increased levels of HERV expression are found, indicating the importance of the regulation of endogenous retroviral elements in the state of pluripotency toward differentiation [10][16]. Interestingly, the type of epigenetic machinery employed appears to be dependent on the evolutionary age of the HERV insertions, as it was demonstrated that DNA methylation occurs mainly on evolutionarily younger insertions, while the histone methylation on the intermediate-age/older ones, with lower CpG density [17].
2.1. DNA Methylation
DNA methyltransferases (DNMTs) drive cytosine methylation. In mammals, three DNTM classes (DNMT1, DNMT3A, and DNMT3B) have been extensively evaluated. Studies in mice indicate the importance of the DNMT3A and DNMT3B function in gametogenesis and early embryonic developmental stages, while low expression of these enzymes characterizes the somatic cells, rendering DNMT-mediated methylation patterns necessary for the orchestration of the epigenetic hallmarks of pluripotency and differentiation. DNMT3L, despite being a catalytically inactive homolog of DNMT3, seems to play an ancillary role in the DNMT3 machinery [18]. Regarding DNMT1, it selectively silences retrotransposon sequences in the developing mouse early embryo genomic (four- and eight-cell stage), mainly in the silencing of the LINE1 (long interspersed nuclear elements) and ERV-K sequences, while in the DNMT1-knockdown mouse embryo, enrichment of the corresponding transcripts was identified, while DNMT1 appears to be non-redundant in the process of embryogenesis [19].
2.2. Histone Methylation
Zinc-finger proteins (ZFPs): Kruppel-associated box (KRAB) zinc-finger proteins (KRAB-ZFPs) are crucial in the regulation of gene expression in mammals and are structurally characterized by two domains, the N-terminal KRAB domain and the C-terminal C2H2 zinc-finger domain, with the former functioning through the recruitment of other cellular transcription factors and the latter through binding to specific sequences for the regulation of transcription. KRAB-ZFPs appear to exert an important role in the silencing of transposable elements (TEs), including HERVs [20] and the nearby genes, through the regulation of the TE-embedded enhancers and promoters [21][22]. The KRAB domain recruits the transcription factor TRIM28 (tripartite motif containing 28 or KRAB-associated protein 1-KAP1), which functions as a scaffold for histone deacetylation proteins heterochromatin protein 1 (HP1) and histone-lysine N-methyltransferase (SETDB1) to control transcription [23], which appears to be pivotal for TE silencing in early embryos [24]. Interestingly, the co-function of TRIM28 and SETDB1 seems to enhance the transcriptional repression of HERVs in a distinct manner, more intensely than each of those factors alone [18]. KRAB-TRIM28 also regulates the expression of long-distance sequences through the extension of heterochromatin, as TRIM28 binds to the 3′ end of genes and leads to the extension of H3K9me3 and HP1β toward the 5′ end [25]. The interactions between KRAB and TRIM28 are summarized in Figure 2.
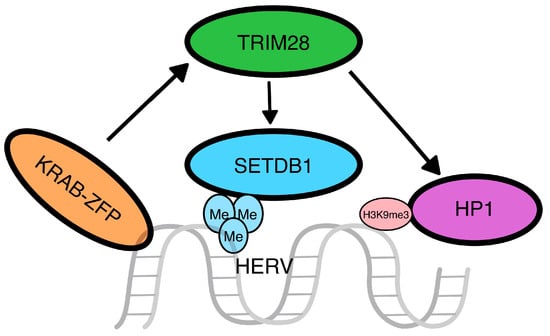
Figure 2. Zinc-finger protein pathway. The Kruppel-associated box zinc-finger proteins (KRAB-ZFP) act as recruiters for other proteins that lead to the silencing of the transposable elements. In detail, the N-term of the KRAB-ZFP recruits a tripartite motif containing 28 (TRIM28), which will work as a scaffold for the SETDB1 and HP1 proteins, a methyltransferase that will methylate the histone H3 on the lysine 9 and an H3K9me3 binding protein, respectively.
Current evidence suggests that ZFPs have been evolutionarily selected to recognize foreign sequences and lead to their silencing. This has been described in the case of ZFP809, a ZFP that appears to demonstrate a specificity in embryonic stem cells, where, despite recognizing a significant number of genomic sites, high-affinity binding and heterochromatin development takes place in primer binding sites (PBSs) associated with HERVs. ZFP809 is elevated during the state of pluripotency compared to somatic cells and, upon ZFP809 depletion in embryonic stem cells, leads to a loss of H3K9me3 and HERV transcriptional release [14]. The importance of the KRAB-ZFP system in the anti-viral protection of the host has been demonstrated in mice with ZFP961, a PBS-Lys binding protein that restrains both endogenous and exogenous retroviruses. The same study identified through chromatin immunoprecipitation sequencing (ChIP-seq analysis) similar PBS-Lys binding proteins in humans, ZNF417 and ZNF587, which, aside from silencing HERVs through facilitating their methylation, appear to restrain the HIV viral infectivity by interfering with viral transcription and integration [26]. The importance of the ZFP HERV transcription control is further shown by the HERV-T and HERV-S TRIM28-mediated silencing and ZNF genes, which appear to participate in innate immunity regulation. They were found to retain their methylation-inducing function in adult human peripheral blood mononuclear cells, where they control interferon responses [27].
Upregulation of the transcription of ZFP genes because of increased HERV element expression in tumors indicates a two-way circuit between HERV transcription and ZFP gene activation. Increased HERV and ZFP expression in these cells was linked to the better prognostic features of these cells, including behavior modifications regarding reduced growth, migrating potential, and invasiveness [28]. Despite the importance of ZFPs and the TRIM28-SETDB1 network in HERV transcriptional control, SETDB1 and H3K9me3 seem to be equally important and independently functioning in the epigenetic silencing of HERVs in differentiated cells [14].
The human silencing hub (HUSH) complex: the HUSH complex is another mechanism that is employed for the defense of the mammalian genome against “invading” sequences, mainly LINE-1 elements and HERVs, through the facilitation of histone H3 lysine 9 trimethylation (H3K9me3) [29]. HUSH has been proposed as a “universal, cell-autonomous genome-surveillance system” and could be considered an innate immunity weapon. HUSH initially recognizes and targets long, intronless transcripts, regarding non-exon organized genome as a conserved hallmark of non-mammalian origin; thus, the transcription of these “foreign” DNA elements is necessary for the H3K9me3 through HUSH [29].
This complex consists of three proteins resident in the nucleus: transcription activation suppressor (TASOR), M-phase phosphoprotein 8 (MPP8), and periphilin [30], with TASOR and periphilin existing in different isoforms as a result of gene conflict [31] (Figure 3). HUSH leads to the activation of MORC family CW-type zinc finger 2 (MORC2), an ATP-dependent chromatin remodeler that compacts chromatin, and SETDB1, for the H3K9me3 of the target sequence [32]. Studies in mice reveal the significant contribution of the HUSH complex to de novo repression of retrotransposons, especially ERVs and LINE-1, with increased specificity to evolutionarily young integrations through TRIM28 activation. Similar findings regarding HUSH silencing of at least LINE-1 elements have been described in humans [33].
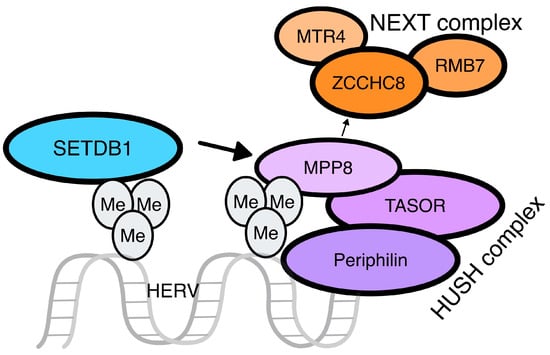
Figure 3. The HUSH complex. The HUSH complex is made up of three different proteins: M-phase phosphoprotein 8 (MPP8), transcription activation suppressor (TASOR), and periphilin. The latest binds on the intronless RNA molecule, while TASOR suppresses its expression. After the stop of the transcription, MPP8 interacts with ZCCHC8, a zinc-finger protein of the nuclear exosome targeting (NEXT) complex, leading to the recruitment of an RNA helicase, namely MTR4, and the RNA-binding protein RBM7. The involvement of the NEXT complex results in the decay of the HERV transcript.
Finally, these two mechanisms (DNA and histone methylation) are not separately and independently functioning but rather interact and synergize during the silencing of genomic regions, especially those of retroviral origin, for the integrity of the host, since in embryonic stem cells in mice, de novo ERV sequence methylation was dependent on the presence of KRAB-ZFPs that could recognize and could properly function on these sequences [34].
3. Histone Acetylation
Acetylation of ε-amino group of lysines is regulated by the histone/lysine acetyltransferase enzymes (HATs/KATs), leading to the more “unstable” chromatin structure and sequences more available to the transcriptional machinery, the effects of which are reversed by histone deacetylases (HDACs) [35]. Interestingly, acetylation can also be applied to non-histone proteins and also be involved in the regulation of different cellular processes, including steps of cell division and differentiation and neuronal function [36]. HDACs, similarly, can induce the deacetylation of both histone and non-histone proteins, leading to an alteration of the transcription in the first case but also to the establishment of other post-translational modifications (PTMs) on the lysines, such as methylation [35], which indicated their mediating role in other regulatory pathways described previously.
Following the consideration of HDAC inhibitors (HDACis) as a means to induce latent HIV-1 provirus expression intended for the eradication of HIV-infected cell reservoirs, interest was drawn to the potential effects of such agents on HERV expression, given their multiple implications in human disease. Results of such studies indicate a rather weak efficacy of acetylation in the control of HERV expression [37][38]. While multiple HERV loci were differentially transcribed following HDACis treatment of HIV(+) primary CD4(+) T-cells, no specific HERV expression pattern was found, such as global HERV upregulation, with the ERV-L family demonstrating a significant downregulation, and the HERV-9 LTR-12 elements were significantly enriched [37]. On the other hand, no substantially different expression of the HERV-K (HML-2) (HK2) env and pol, syncytin-1, and syncytin-2 was found in another study [38].
4. Cytosine Deamination
The cytidine deaminases APOBEC3 (A3) family mediates the deamination as a mechanism of control of HERV expression which functions both on DNA and RNA molecules [39]. This system, through the expression of seven members of the A3 family, is considered a part of the human innate immunity machinery to protect the host against exogenous and endogenous retroviruses [40]. It has been demonstrated that this protein family represses retroelement expression through binding at their genomic sites in the human DNA [39], while A3G exerts its antiretroviral functions through the inhibition of the retroviral reverse transcription, aiming at the control of the retroviral replication [41].
The evolutionary expansion of the APOBEC protein family could be considered a milestone in the human genome, in terms of protection against endogenous pathogens as inferred compared to other mammals such as rodents, mainly through the A3A- and A3B-mediated regulation of LINE-1 elements and HERVs [42]. A3 genes have been amplified in the mammalian genomes in multi-species studies, and this expansion correlated positively to the germline expansion of HERVs; also, this amplification dates concurrently with ancient retroviral invasions. Finally, deamination mediated by the A3 system leads to increased A-to-G point mutation counts in ERV sequences, rendering them more susceptible to deactivation through frameshifts or premature stop codons [42].
5. Chromatin Remodeling
Chromatin remodeling, the histone shuffle to render DNA regions accessible for transcription and/or epigenetic modification, is another HERV silencing mechanism [43]. One of the mediators of such changes is the SWItch-sucrose non-fermentable (SWI-SNF) complex. Polybromo 1 (PBRM1), a member of the SWI-SNF complex, deregulation leads to the deregulated expression of HERV-ERI in clear cell renal carcinoma with a still unknown mechanism. However, repression of hypoxia inducible factor 1α (HIF-1α) seems to be a part of this SWI-SNF-mediated process [44].
Another significant protein family in the epigenetic control of HERVs is the MORC proteins that, after reducing DNA accessibility to the transcriptional machinery, aid H3K9 tri-methylation through the HUSH-complex and DNA methylation by DNMTs [45]. Interestingly, MORC3’s proper function is pivotal for ERV chromatin regulation in mice, as global ERV dysregulation was described in MORC3 knockout mice. The HUSH complex also recruits the nuclear exosome targeting (NEXT) complex as a post-transcriptional means of intronless RNA decay after its formation. HUSH and NEXT are simultaneously recruited in the nucleus, where following their binding, they regulate ERV expression both at a transcriptional and post-transcriptional level [46] (Figure 3).
References
- Mager, D.L.; Stoye, J.P. Mammalian Endogenous Retroviruses. Microbiol. Spectr. 2015, 3, MDNA3-0009-2014.
- Lee, Y.N.; Bieniasz, P.D. Reconstitution of an Infectious Human Endogenous Retrovirus. PLoS Pathog. 2007, 3, e10.
- Marchi, E.; Kanapin, A.; Magiorkinis, G.; Belshaw, R. Unfixed Endogenous Retroviral Insertions in the Human Population. J. Virol. 2014, 88, 9529–9537.
- Mao, J.; Zhang, Q.; Cong, Y.S. Human Endogenous Retroviruses in Development and Disease. Comput. Struct. Biotechnol. J. 2021, 19, 5978–5986.
- Vargiu, L.; Rodriguez-Tomé, P.; Sperber, G.O.; Cadeddu, M.; Grandi, N.; Blikstad, V.; Tramontano, E.; Blomberg, J. Classification and Characterization of Human Endogenous Retroviruses; Mosaic Forms Are Common. Retrovirology 2016, 13, 7.
- Blomberg, J.; Benachenhou, F.; Blikstad, V.; Sperber, G.; Mayer, J. Classification and Nomenclature of Endogenous Retroviral Sequences (ERVs): Problems and Recommendations. Gene 2009, 448, 115–123.
- Fauquet, C.; Fargette, D. International Committee on Taxonomy of Viruses and the 3,142 Unassigned Species. Virol. J. 2005, 2, 64.
- Seifarth, W.; Frank, O.; Zeilfelder, U.; Spiess, B.; Greenwood, A.D.; Hehlmann, R.; Leib-Mösch, C. Comprehensive Analysis of Human Endogenous Retrovirus Transcriptional Activity in Human Tissues with a Retrovirus-Specific Microarray. J. Virol. 2005, 79, 341–352.
- Grandi, N.; Cadeddu, M.; Blomberg, J.; Tramontano, E. Contribution of Type W Human Endogenous Retroviruses to the Human Genome: Characterization of HERV-W Proviral Insertions and Processed Pseudogenes. Retrovirology 2016, 13, 67.
- Göke, J.; Lu, X.; Chan, Y.-S.; Ng, H.-H.; Ly, L.-H.; Sachs, F.; Szczerbinska, I. Dynamic Transcription of Distinct Classes of Endogenous Retroviral Elements Marks Specific Populations of Early Human Embryonic Cells. Cell Stem Cell 2015, 16, 135–141.
- Ito, J.; Sugimoto, R.; Nakaoka, H.; Yamada, S.; Kimura, T.; Hayano, T.; Inoue, I. Systematic Identification and Characterization of Regulatory Elements Derived from Human Endogenous Retroviruses. PLoS Genet. 2017, 13, e1006883.
- Medstrand, P.; Landry, J.-R.; Mager, D.L. Long Terminal Repeats Are Used as Alternative Promoters for the Endothelin B Receptor and Apolipoprotein C-I Genes in Humans. J. Biol. Chem. 2001, 276, 1896–1903.
- Jern, P.; Coffin, J.M. Effects of Retroviruses on Host Genome Function. Annu. Rev. Genet. 2008, 42, 709–732.
- Wolf, G.; Yang, P.; Füchtbauer, A.C.; Füchtbauer, E.-M.; Silva, A.M.; Park, C.; Wu, W.; Nielsen, A.L.; Pedersen, F.S.; Macfarlan, T.S. The KRAB Zinc Finger Protein ZFP809 Is Required to Initiate Epigenetic Silencing of Endogenous Retroviruses. Genes Dev. 2015, 29, 538–554.
- Feldman, N.; Gerson, A.; Fang, J.; Li, E.; Zhang, Y.; Shinkai, Y.; Cedar, H.; Bergman, Y. G9a-Mediated Irreversible Epigenetic Inactivation of Oct-3/4 during Early Embryogenesis. Nat. Cell Biol. 2006, 8, 188–194.
- Khodosevich, K. Large-Scale Determination of the Methylation Status of Retrotransposons in Different Tissues Using a Methylation Tags Approach. Nucleic Acids Res. 2004, 32, 31e.
- Ohtani, H.; Liu, M.; Zhou, W.; Liang, G.; Jones, P.A. Switching Roles for DNA and Histone Methylation Depend on Evolutionary Ages of Human Endogenous Retroviruses. Genome Res. 2018, 28, 1147–1157.
- Geis, F.K.; Goff, S.P. Silencing and Transcriptional Regulation of Endogenous Retroviruses: An Overview. Viruses 2020, 12, 884.
- Min, B.; Park, J.S.; Jeong, Y.S.; Jeon, K.; Kang, Y.-K. Dnmt1 Binds and Represses Genomic Retroelements via DNA Methylation in Mouse Early Embryos. Nucleic Acids Res. 2020, 48, 8431–8444.
- Peng, H.; Begg, G.E.; Harper, S.L.; Friedman, J.R.; Speicher, D.W.; Rauscher, F.J. Biochemical Analysis of the Kruppel-Associated Box (KRAB) Transcriptional Repression Domain. J. Biol. Chem. 2000, 275, 18000–18010.
- Schön, U.; Diem, O.; Leitner, L.; Gönzburg, W.H.; Mager, D.L.; Salmons, B.; Leib-Mösch, C. Human Endogenous Retroviral Long Terminal Repeat Sequences as Cell Type-Specific Promoters in Retroviral Vectors. J. Virol. 2009, 83, 12643–12650.
- Ruda, V.M.; Akopov, S.B.; Trubetskoy, D.O.; Manuylov, N.L.; Vetchinova, A.S.; Zavalova, L.L.; Nikolaev, L.G.; Sverdlov, E.D. Tissue Specificity of Enhancer and Promoter Activities of a HERV-K(HML-2) LTR. Virus Res. 2004, 104, 11–16.
- Turelli, P.; Castro-Diaz, N.; Marzetta, F.; Kapopoulou, A.; Raclot, C.; Duc, J.; Tieng, V.; Quenneville, S.; Trono, D. Interplay of TRIM28 and DNA Methylation in Controlling Human Endogenous Retroelements. Genome Res. 2014, 24, 1260–1270.
- Wolf, D.; Goff, S.P. TRIM28 Mediates Primer Binding Site-Targeted Silencing of Murine Leukemia Virus in Embryonic Cells. Cell 2007, 131, 46–57.
- Groner, A.C.; Meylan, S.; Ciuffi, A.; Zangger, N.; Ambrosini, G.; Dénervaud, N.; Bucher, P.; Trono, D. KRAB–Zinc Finger Proteins and KAP1 Can Mediate Long-Range Transcriptional Repression through Heterochromatin Spreading. PLoS Genet. 2010, 6, e1000869.
- Yang, B.; Fang, L.; Gao, Q.; Xu, C.; Xu, J.; Chen, Z.-X.; Wang, Y.; Yang, P. Species-Specific KRAB-ZFPs Function as Repressors of Retroviruses by Targeting PBS Regions. Proc. Natl. Acad. Sci. USA 2022, 119, e2119415119.
- Tie, C.H.; Fernandes, L.; Conde, L.; Robbez-Masson, L.; Sumner, R.P.; Peacock, T.; Rodriguez-Plata, M.T.; Mickute, G.; Gifford, R.; Towers, G.J.; et al. KAP1 Regulates Endogenous Retroviruses in Adult Human Cells and Contributes to Innate Immune Control. EMBO Rep. 2018, 19, e45000.
- Ito, J.; Kimura, I.; Soper, A.; Coudray, A.; Koyanagi, Y.; Nakaoka, H.; Inoue, I.; Turelli, P.; Trono, D.; Sato, K. Endogenous Retroviruses Drive KRAB Zinc-Finger Protein Family Expression for Tumor Suppression. Sci. Adv. 2020, 6, eabc3020.
- Seczynska, M.; Bloor, S.; Cuesta, S.M.; Lehner, P.J. Genome Surveillance by HUSH-Mediated Silencing of Intronless Mobile Elements. Nature 2022, 601, 440–445.
- Tchasovnikarova, I.A.; Timms, R.T.; Matheson, N.J.; Wals, K.; Antrobus, R.; Göttgens, B.; Dougan, G.; Dawson, M.A.; Lehner, P.J. Epigenetic Silencing by the HUSH Complex Mediates Position-Effect Variegation in Human Cells. Science 2015, 348, 1481–1485.
- Chougui, G.; Munir-Matloob, S.; Matkovic, R.; Martin, M.M.; Morel, M.; Lahouassa, H.; Leduc, M.; Ramirez, B.C.; Etienne, L.; Margottin-Goguet, F. HIV-2/SIV Viral Protein X Counteracts HUSH Repressor Complex. Nat. Microbiol. 2018, 3, 891–897.
- Seczynska, M.; Lehner, P.J. The Sound of Silence: Mechanisms and Implications of HUSH Complex Function. Trends Genet. 2023, 39, 251–267.
- Robbez-Masson, L.; Tie, C.H.C.; Conde, L.; Tunbak, H.; Husovsky, C.; Tchasovnikarova, I.A.; Timms, R.T.; Herrero, J.; Lehner, P.J.; Rowe, H.M. The HUSH Complex Cooperates with TRIM28 to Repress Young Retrotransposons and New Genes. Genome Res. 2018, 28, 836–845.
- Rowe, H.M.; Friedli, M.; Offner, S.; Verp, S.; Mesnard, D.; Marquis, J.; Aktas, T.; Trono, D. De Novo DNA Methylation of Endogenous Retroviruses Is Shaped by KRAB-ZFPs/KAP1 and ESET. Development 2013, 140, 519–529.
- Seto, E.; Yoshida, M. Erasers of Histone Acetylation: The Histone Deacetylase Enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713.
- Shvedunova, M.; Akhtar, A. Modulation of Cellular Processes by Histone and Non-Histone Protein Acetylation. Nat. Rev. Mol. Cell Biol. 2022, 23, 329–349.
- Hurst, T.; Pace, M.; Katzourakis, A.; Phillips, R.; Klenerman, P.; Frater, J.; Magiorkinis, G. Human Endogenous Retrovirus (HERV) Expression Is Not Induced by Treatment with the Histone Deacetylase (HDAC) Inhibitors in Cellular Models of HIV-1 Latency. Retrovirology 2016, 13, 10.
- White, C.H.; Beliakova-Bethell, N.; Lada, S.M.; Breen, M.S.; Hurst, T.P.; Spina, C.A.; Richman, D.D.; Frater, J.; Magiorkinis, G.; Woelk, C.H. Transcriptional Modulation of Human Endogenous Retroviruses in Primary CD4+ T Cells Following Vorinostat Treatment. Front. Immunol. 2018, 9, 603.
- Esnault, C.; Priet, S.; Ribet, D.; Heidmann, O.; Heidmann, T. Restriction by APOBEC3 Proteins of Endogenous Retroviruses with an Extracellular Life Cycle: Ex Vivo Effects and in Vivo “Traces” on the Murine IAPE and Human HERV-K Elements. Retrovirology 2008, 5, 75.
- Delviks-Frankenberry, K.A.; Desimmie, B.A.; Pathak, V.K. Structural Insights into APOBEC3-Mediated Lentiviral Restriction. Viruses 2020, 12, 587.
- Chiu, Y.-L.; Greene, W.C. The APOBEC3 Cytidine Deaminases: An Innate Defensive Network Opposing Exogenous Retroviruses and Endogenous Retroelements. Annu. Rev. Immunol. 2008, 26, 317–353.
- Ito, J.; Gifford, R.J.; Sato, K. Retroviruses Drive the Rapid Evolution of Mammalian APOBEC3 Genes. Proc. Natl. Acad. Sci. USA 2020, 117, 610–618.
- Wilson, B.G.; Roberts, C.W.M. SWI/SNF Nucleosome Remodellers and Cancer. Nat. Rev. Cancer 2011, 11, 481–492.
- Zhou, M.; Leung, J.Y.; Gessner, K.H.; Hepperla, A.J.; Simon, J.M.; Davis, I.J.; Kim, W.Y. PBRM1 Inactivation Promotes Upregulation of Human Endogenous Retroviruses in a HIF-Dependent Manner. Cancer Immunol. Res. 2022, 10, 285–290.
- Groh, S.; Milton, A.V.; Marinelli, L.K.; Sickinger, C.V.; Russo, A.; Bollig, H.; de Almeida, G.P.; Schmidt, A.; Forné, I.; Imhof, A.; et al. Morc3 Silences Endogenous Retroviruses by Enabling Daxx-Mediated Histone H3.3 Incorporation. Nat. Commun. 2021, 12, 5996.
- Garland, W.; Müller, I.; Wu, M.; Schmid, M.; Imamura, K.; Rib, L.; Sandelin, A.; Helin, K.; Jensen, T.H. Chromatin Modifier HUSH Co-Operates with RNA Decay Factor NEXT to Restrict Transposable Element Expression. Mol. Cell 2022, 82, 1691–1707.e8.
More
Information
Subjects:
Virology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
680
Revisions:
2 times
(View History)
Update Date:
26 Feb 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





