Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Hegias Mira Bontenbal | -- | 2098 | 2024-02-22 12:47:08 | | | |
| 2 | Peter Tang | + 1 word(s) | 2099 | 2024-02-23 07:01:32 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Luchsinger-Morcelle, S.J.; Gribnau, J.; Mira-Bontenbal, H. DNA Elements That Impact Xist Expression. Encyclopedia. Available online: https://encyclopedia.pub/entry/55350 (accessed on 07 February 2026).
Luchsinger-Morcelle SJ, Gribnau J, Mira-Bontenbal H. DNA Elements That Impact Xist Expression. Encyclopedia. Available at: https://encyclopedia.pub/entry/55350. Accessed February 07, 2026.
Luchsinger-Morcelle, Samuel Jesus, Joost Gribnau, Hegias Mira-Bontenbal. "DNA Elements That Impact Xist Expression" Encyclopedia, https://encyclopedia.pub/entry/55350 (accessed February 07, 2026).
Luchsinger-Morcelle, S.J., Gribnau, J., & Mira-Bontenbal, H. (2024, February 22). DNA Elements That Impact Xist Expression. In Encyclopedia. https://encyclopedia.pub/entry/55350
Luchsinger-Morcelle, Samuel Jesus, et al. "DNA Elements That Impact Xist Expression." Encyclopedia. Web. 22 February, 2024.
Copy Citation
Compensation for the gene dosage disequilibrium between sex chromosomes in mammals is achieved in female cells by repressing one of its X chromosomes through a process called X chromosome inactivation (XCI), exemplifying the control of gene expression by epigenetic mechanisms. A critical player in this mechanism is Xist, a long, non-coding RNA upregulated from a single X chromosome during early embryonic development in female cells. Xist is regulated at different levels in cis and trans, such as DNA elements, transcription factors, other regulatory proteins, long non-coding RNAs and the chromatin and topological landscape surrounding Xist. This entry focuses on DNA elements that govern Xist expression.
X chromosome inactivation
Xist
epigenetics
1. Introduction
Dosage compensation of gene expression between chromosomes is essential for survival. Female mammalian cells carry two X chromosomes, while male cells carry an X and a Y chromosome, generating an X-linked gene dosage imbalance between the sexes. To achieve dosage compensation, female diploid epiblast cells inactivate a single X chromosome very early during embryonic development via a complex process known as X chromosome inactivation (XCI). This process results in the epigenetic silencing of one randomly selected X chromosome (Xi), indicating that female and male cells must sense how many X chromosomes they carry.
In mice, two waves of epigenetic silencing occur. Firstly, imprinted XCI (iXCI) leads to the inactivation of the paternal X chromosome following fertilization. This paternal Xi is later re-activated in the inner cell mass of the embryo, and a second, random XCI (rXCI) wave occurs, where a single X is randomly chosen for inactivation in the early epiblast. Most in vitro studies of XCI are performed in mouse embryonic stem cells (ESCs), which carry two active X chromosomes (Xa) and initiate rXCI upon exit of pluripotency and differentiation, recapitulating events in the epiblast of embryos. However, not all genes on the Xi are silenced; they are called escapees or escaping genes. Even though XCI happens in all mammals, different species show different patterns of it. While mice display both types of XCI, rabbits, monkeys, and humans, for instance, only show rXCI, and marsupials display iXCI only. This illustrates the variety of XCI mechanisms present within the mammalian class. New data are emerging on different mechanisms in other mammals, such as rabbits, monkeys and especially humans (reviewed in [1]). However, since most XCI research has been performed in mice and very exciting new data are still being generated nowadays, the researchers focus this entry on mouse rXCI.
1.1. Kicking off X Chromosome Inactivation: The Stochastic Model
In 1971, Mary Lyon proposed a model where a diffusible X-linked factor would guarantee XCI of a single X [2]. This model has been further developed into a stochastic model, where female cells sense X chromosome dosage, leading to the inactivation of a single X chromosome while preventing inactivation of the single X chromosome in male cells. The stochastic model proposes that XCI is achieved by a tightly controlled balance between X-encoded activators and autosomally encoded repressors [3]. Many autosomally encoded repressors are pluripotency factors or are linked to the pluripotent state and therefore prevent the inactivation of an X chromosome in the inner cell mass of the embryo (ICM) or in ESCs. ESCs are derived from the ICM, which will give rise to the embryo proper. Upon differentiation, the downregulation of repressors of XCI and upregulation of activators of XCI would tilt the balance towards XCI. Thus, female exclusive activation of XCI is mediated by the double dose of X-encoded XCI activators that are required to overcome the threshold set by the XCI repressors. Initiation of XCI is stochastic, and a negative feedback loop involving rapid silencing of some XCI activators prevents inactivation of the second X chromosome. The application of novel machine learning and mathematical modeling techniques indicates that XCI dynamics can be recapitulated based on the principles of the stochastic model with a limited number of activators and repressors regulating XCI [4][5].
1.2. The X Inactivation Center and the Tsix/Xist Tandem
Research on truncated human X chromosomes and balanced X-autosome translocations led to the discovery of the X inactivation center (Xic), a locus on the X chromosome essential for XCI [6][7]. The mouse Xic spans many long non-coding RNAs and several protein-coding genes, covering at least 800kb (reviewed in [8]). Later, transgenic studies showed the long non-coding RNA (lncRNA) gene Xist, embedded within the Xic, to be required for XCI to happen. Xist is critical for both iXCI and rXCI in mice. Endogenous Xist deletions prevent XCI from happening on the X chromosome carrying the deletion, while ectopic insertions on autosomes result in their silencing [9][10]. The mouse Xist gene is a 22 kb-long gene with no conserved open reading frame (ORF) and is transcribed, polyadenylated, and alternatively spliced into a 15 kb-long lncRNA [11][12][13]. Xist is transcribed antisense to another lncRNA named Tsix that spans Xist entirely and negatively regulates its expression during development [14]. Biallelic expression of Tsix in the pluripotent state thus maintains the silent state of Xist in female ESCs. Exit of pluripotency triggers a break from symmetric Xist expression to an asymmetric Tsix/Xist expression state, where Tsix expression is maintained on the Xa while Xist expression is greatly upregulated on the selected Xi. Finally, Xist expression is locked on the Xi, and the eventual downregulation of Tsix from the Xa happens once XCI is established (Figure 1). Many of these types of antisense gene expression regulation have been described in animals and plants, such as imprinted genes or the Flc-COOLAIR gene-tandem involved in plant flowering [15].
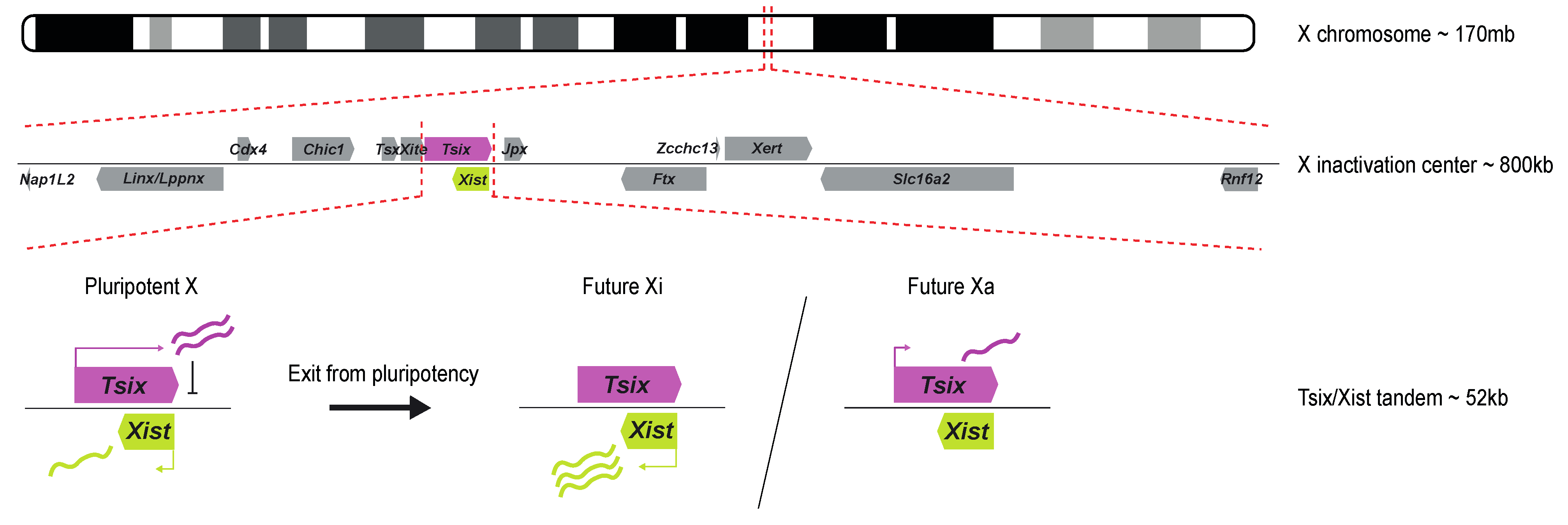
Figure 1. Schematic overview of the X inactivation center and the Tsix/Xist tandem. The Xic is located at ~103 Mb on the mouse X chromosome (mm10). It contains several lncRNAs, such as Linx/Lppnx, Tsx, Xite, Tsix, Xist, Jpx, Ftx, and Xert, and several coding genes, such as Nap1L2, Cdx4, Chic1, Slc16a2, and Rnf12. Xist and Tsix are two antisense genes at the center of the Xic. In the pluripotent state, their expression is symmetric between both X chromosomes: Tsix is biallelically expressed and represses Xist, leading to low levels of Xist expression. Upon XCI, Tsix is downregulated from the future Xi, resulting in monoallelic Xist upregulation, while the future Xa maintains low levels of Tsix, further suppressing Xist expression. The symmetry is broken, and XCI happens on a single chromosome in diploid female cells.
2. The Promoter Region of Xist
The discovery of the Xist gene was a significant breakthrough in understanding the mechanism of XCI. Its lncRNA product is now accepted as the master regulator behind the epigenetic silencing of one of the two X chromosomes in female mammals. Xist was first identified and isolated from a mouse cDNA library in 1991 [7]. It was discovered that Xist RNA spreads in cis, interacting with the chromatin of the X chromosome, leading to the spread of heterochromatinization and subsequent inactivation of the chromosome [12][16]. In an effort to understand the transcriptional regulation of Xist and thus the mechanisms kicking off XCI, follow-up studies centered on the characterization of the Xist promoter.
Early studies identified the mouse Xist gene’s minimal promoter region to be approximately 0.4 kb in size and located right upstream of the major transcriptional start site (TSS). This minimal promoter is a weak constitutive TATA-like promoter with a possible initiator element spanning the TSS [17]. Xist promoter sequences have been compared between several mammals, including humans, mice, rabbits, and horses, showing a high degree of conservation between these species [18]. Follow-up studies identified multiple Xist promoters, termed P1, P2, and P0, with P1 being the first identified region just upstream of Xist’s TSS. The P0 promoter of Xist was mapped 6.5 kb upstream of the TSS and gives rise to unstable Xist transcripts. The P2 promoter was mapped 1.5 kb downstream of the TSS and thus within Xist exon 1, giving rise to a stable Xist transcript (Figure 2A,B). Promoter switching from P0 to P1/P2 was then proposed as a regulatory mechanism controlling Xist expression with the exit of pluripotency [19]. However, recent chromatin immunoprecipitation assays followed by sequencing (ChIP-seq) of the P2 promoter of Xist seem to indicate this region might act as an internal regulatory element, or enhancer, rather than a promoter [20][21].
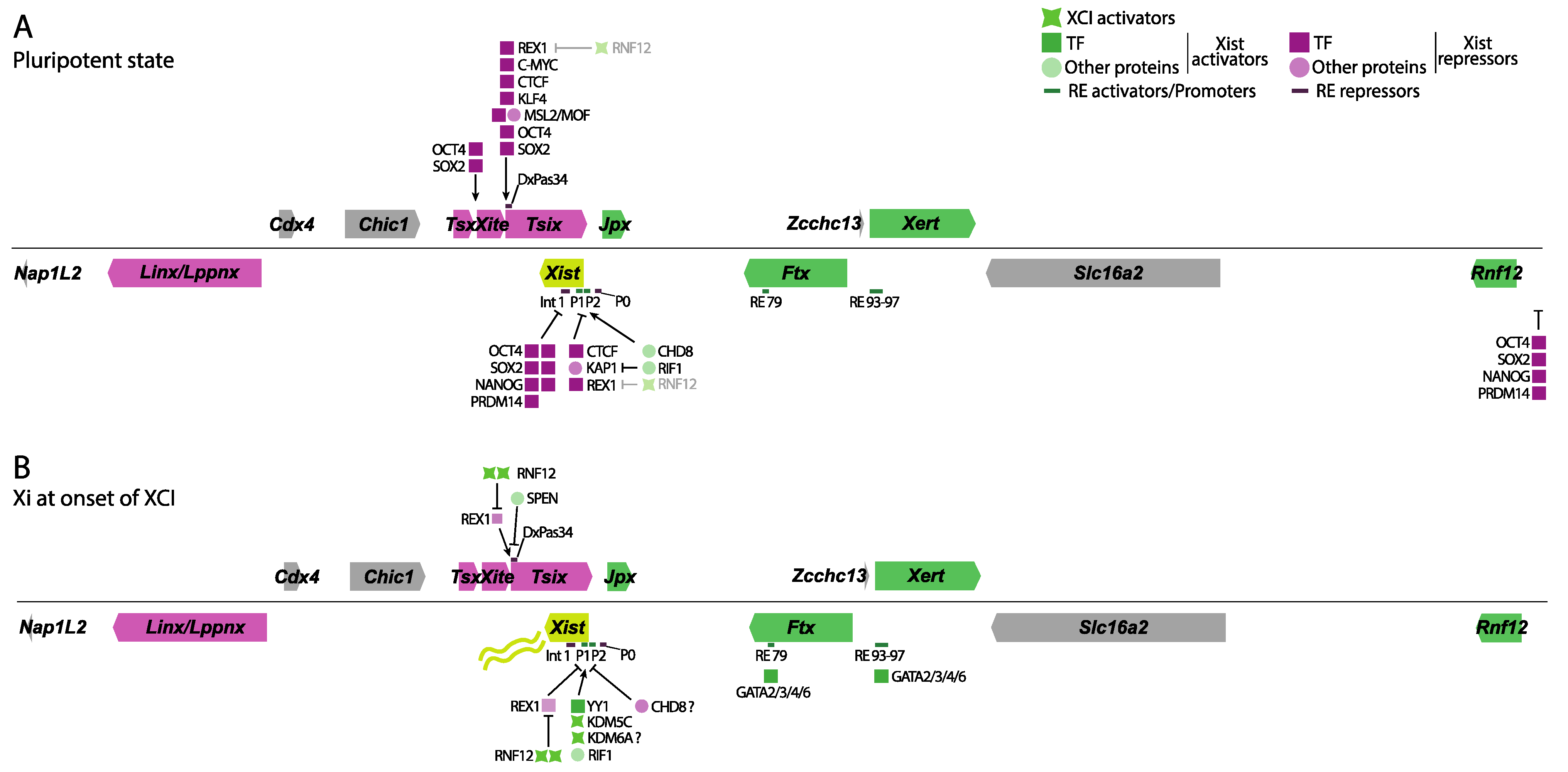
Figure 2. Overview of the different proteins and REs involved in Xist and Tsix regulation. Genes that have an activating direct or indirect role on Xist are shown in green, while genes involved in Xist repression are shown in magenta. Several TF, other proteins, XCI activators, and RE are shown in green or magenta based on their effect on Xist expression. (A) In pluripotency, the pluripotency factor network (OCT4, NANOG, SOX2, REX1, PRDM14) keeps Tsix active and represses Xist, either directly by binding to Intron 1 or its promoter, or indirectly through Tsix or by inhibiting the Xist activator RNF12. CTCF and KAP1 also work towards inhibiting its expression, while the MSL complex supports Tsix expression from its promoter. Another set of activating proteins, such as CHD8 and RIF1, supports low levels of Xist expression. (B) At the onset of XCI, reduced pluripotency factor concentrations lead to decreased Tsix expression. Reduction in REX1 is further aided by increased RNF12 expression due to the disappearance of its pluripotent repressors. SPEN is required to shut down Tsix expression to allow Xist upregulation. YY1, the paralog of REX1, binds to the P2 promoter of Xist, activating it. KDM5C and KDM6A also seem to bind there, leading to the demethylation of H3K4me2/3 to H3K4me1, a mark of enhancers, while seemingly removing H3K27me3. CHD8, however, seems to have an opposing role at the onset of XCI during differentiation because it decreases YY1s accessibility to Xist’s promoter. CHD8 seems to fine-tune Xist expression depending on the developmental context. Several of the GATA binding factors are required for Xist expression during differentiation by binding several of its regulatory sequences. Shades of magenta show Xist repressors, while shades of green indicate Xist activators and XCI activators.
3. Distant Regulatory Regions
While the promoter regions of Xist drive its expression, distant DNA regulatory elements (RE) seem to play a role in titrating its expression. A regulatory region located ∼10 kb downstream of Xist TSS, and thus within Xist intron 1, was shown to bind several pluripotency factors and play a role in Xist repression in the pluripotent state (Figure 2A) [22]. A subsequent study confirmed these findings through genetic deletions of Xist intron 1. Transgenic male ESCs carrying an Xist intron 1 deletion show moderately upregulated Xist expression in the pluripotent state, which is exacerbated upon Tsix co-removal [23]. However, in two contrasting studies, deletion of Xist intron 1 in female ES cells does not seem to impact Xist expression both in vivo and in vitro but rather skews the future Xi choice towards the mutated allele [24][25].
Identification of REs involved in Xist expression has proved to be a challenging task due to numerous factors influencing Xist expression within the Xic. Distinguishing direct from indirect effects on Xist expression via genetic perturbations within the Xic is difficult. However, a recent publication has successfully addressed this challenge. Using an elegant screening approach taking advantage of dCas9 fused to the repressor KRAB (CRISPRi) and detecting Xist levels by fluorescently activated cell sorting, several candidate regions were epigenetically and systematically silenced [26]. As expected, epigenetic inactivation of the previously discussed Xist promoter regions, as well as of known Xist regulators, impairs Xist upregulation upon exit of pluripotency, serving as a validation of the screening approach. A novel regulatory cluster located ~150 kb upstream of the Xist TSS, termed RE93-97, was also identified. Targeting this cluster with CRISPRi revealed a gradual decrease in Xist expression, pointing to RE93-97’s role as a Xist enhancer cluster. Furthermore, capture Hi-C analysis revealed RE93-97 interacts with Xist’s P2 promoter upon exit of pluripotency, suggesting this regulatory cluster responds to differentiation cues.
In a follow-up study by the same group, another pivotal RE responsible for driving Xist expression was identified [27]. Employing a CRISPR activation screening approach, several protein factors that result in Xist expression upon their overexpression were identified. A comprehensive analysis of potential candidates and their binding sites revealed that the GATA family of transcription factors binds not only RE93-97 but also a newly discovered regulatory element located ~100 kb upstream of the Xist TSS termed RE79. Genetic deletions of RE97, as well as RE79, validated previous discoveries regarding the significance of RE93-97 in rXCI while elucidating the role of RE79 in driving Xist upregulation expression in iXCI.
Gene transcription regulation is achieved not only by many different protein factors binding to promoters and REs, such as RNA Pol II, transcription factors (TFs), chromatin remodellers, etc., but also through complex chromatin architecture.
References
- Khan, S.A.; Theunissen, T.W. Modeling X-Chromosome Inactivation and Reactivation during Human Development. Curr. Opin. Genet. Dev. 2023, 82, 102096.
- Lyon, M.F. Possible Mechanisms of X Chromosome Inactivation. Nat. New Biol. 1971, 232, 229–232.
- Monkhorst, K.; Jonkers, I.; Rentmeester, E.; Grosveld, F.; Gribnau, J. X Inactivation Counting and Choice Is a Stochastic Process: Evidence for Involvement of an X-Linked Activator. Cell 2008, 132, 410–421.
- Mutzel, V.; Okamoto, I.; Dunkel, I.; Saitou, M.; Giorgetti, L.; Heard, E.; Schulz, E.G. A Symmetric Toggle Switch Explains the Onset of Random X Inactivation in Different Mammals. Nat. Struct. Mol. Biol. 2019, 26, 350–360.
- De Andrade e Sousa, L.B.; Jonkers, I.; Syx, L.; Dunkel, I.; Chaumeil, J.; Picard, C.; Foret, B.; Chen, C.-J.; Lis, J.T.; Heard, E.; et al. Kinetics of Xist-Induced Gene Silencing Can Be Predicted from Combinations of Epigenetic and Genomic Features. Genome Res. 2019, 29, 1087–1099.
- Rastan, S.; Robertson, E.J. X-Chromosome Deletions in Embryo-Derived (EK) Cell Lines Associated with Lack of X-Chromosome Inactivation. Development 1985, 90, 379–388.
- Brown, C.J.; Ballabio, A.; Rupert, J.L.; Lafreniere, R.G.; Grompe, M.; Tonlorenzi, R.; Willard, H.F. A Gene from the Region of the Human X Inactivation Centre Is Expressed Exclusively from the Inactive X Chromosome. Nature 1991, 349, 38–44.
- Furlan, G.; Galupa, R. Mechanisms of Choice in X-Chromosome Inactivation. Cells 2022, 11, 535.
- Heard, E.; Mongelard, F.; Arnaud, D.; Chureau, C.; Vourc’h, C.; Avner, P. Human XIST Yeast Artificial Chromosome Transgenes Show Partial X Inactivation Center Function in Mouse Embryonic Stem Cells. Proc. Natl. Acad. Sci. USA 1999, 96, 6841–6846.
- Penny, G.D.; Kay, G.F.; Sheardown, S.A.; Rastan, S.; Brockdorff, N. Requirement for Xist in X Chromosome Inactivation. Nature 1996, 379, 131–137.
- Memili, E.; Hong, Y.-K.; Kim, D.-H.; Ontiveros, S.D.; Strauss, W.M. Murine Xist RNA Isoforms Are Different at Their 3′ Ends: A Role for Differential Polyadenylation. Gene 2001, 266, 131–137.
- Brockdorff, N.; Ashworth, A.; Kay, G.F.; McCabe, V.M.; Norris, D.P.; Cooper, P.J.; Swift, S.; Rastan, S. The Product of the Mouse Xist Gene Is a 15 Kb Inactive X-Specific Transcript Containing No Conserved ORF and Located in the Nucleus. Cell 1992, 71, 515–526.
- Yue, M.; Ogawa, Y. CRISPR/Cas9-Mediated Modulation of Splicing Efficiency Reveals Short Splicing Isoform of Xist RNA Is Sufficient to Induce X-Chromosome Inactivation. Nucleic Acids Res. 2018, 46, e26.
- Lee, J.T.; Lu, N. Targeted Mutagenesis of Tsix Leads to Nonrandom X Inactivation. Cell 1999, 99, 47–57.
- Rosa, S.; Duncan, S.; Dean, C. Mutually Exclusive Sense–Antisense Transcription at FLC Facilitates Environmentally Induced Gene Repression. Nat. Commun. 2016, 7, 13031.
- Csankovszki, G.; Panning, B.; Bates, B.; Pehrson, J.R.; Jaenisch, R. Conditional Deletion of Xist Disrupts Histone MacroH2A Localization but Not Maintenance of X Inactivation. Nat. Genet. 1999, 22, 323–324.
- Sheardown, S.A.; Duthie, S.M.; Johnston, C.M.; Newall, A.E.; Formstone, E.J.; Arkell, R.M.; Nesterova, T.B.; Alghisi, G.C.; Rastan, S.; Brockdorff, N. Stabilization of Xist RNA Mediates Initiation of X Chromosome Inactivation. Cell 1997, 91, 99–107.
- Hendrich, B.D.; Plenge, R.M.; Willard, H.F. Identification and Characterization of the Human XIST Gene Promoter: Implications for Models of X Chromosome Inactivation. Nucleic Acids Res. 1997, 25, 2661–2671.
- Johnston, C.M.; Nesterova, T.B.; Formstone, E.J.; Newall, A.E.T.; Duthie, S.M.; Sheardown, S.A.; Brockdorff, N. Developmentally Regulated Xist Promoter Switch Mediates Initiation of X Inactivation. Cell 1998, 94, 809–817.
- Makhlouf, M.; Ouimette, J.F.; Oldfield, A.; Navarro, P.; Neuillet, D.; Rougeulle, C. A Prominent and Conserved Role for YY1 in Xist Transcriptional Activation. Nat. Commun. 2014, 5, 4878.
- Samanta, M.K.; Gayen, S.; Harris, C.; Maclary, E.; Murata-Nakamura, Y.; Malcore, R.M.; Porter, R.S.; Garay, P.M.; Vallianatos, C.N.; Samollow, P.B.; et al. Activation of Xist by an Evolutionarily Conserved Function of KDM5C Demethylase. Nat Commun 2022, 13, 2602.
- Navarro, P.; Chambers, I.; Karwacki-Neisius, V.; Chureau, C.; Morey, C.; Rougeulle, C.; Avner, P. Molecular Coupling of Xist Regulation and Pluripotency. Science 2008, 321, 1693–1695.
- Nesterova, T.B.; Senner, C.E.; Schneider, J.; Alcayna-Stevens, T.; Tattermusch, A.; Hemberger, M.; Brockdorff, N. Pluripotency Factor Binding and Tsix Expression Act Synergistically to Repress Xist in Undifferentiated Embryonic Stem Cells. Epigenetics Chromatin 2011, 4, 17.
- Barakat, T.S.; Gunhanlar, N.; Pardo, C.G.; Achame, E.M.; Ghazvini, M.; Boers, R.; Kenter, A.; Rentmeester, E.; Grootegoed, J.A.; Gribnau, J. RNF12 Activates Xist and Is Essential for X Chromosome Inactivation. PLoS Genet. 2011, 7, e1002001.
- Minkovsky, A.; Barakat, T.S.; Sellami, N.; Chin, M.H.; Gunhanlar, N.; Gribnau, J.; Plath, K. The Pluripotency Factor-Bound Intron 1 of Xist Is Dispensable for X Chromosome Inactivation and Reactivation In Vitro and In Vivo. Cell Rep. 2013, 3, 905–918.
- Gjaltema, R.A.F.; Schwämmle, T.; Kautz, P.; Robson, M.; Schöpflin, R.; Lustig, L.R.; Brandenburg, L.; Dunkel, I.; Vechiatto, C.; Ntini, E.; et al. Distal and Proximal Cis-Regulatory Elements Sense X Chromosome Dosage and Developmental State at the Xist Locus. Mol. Cell 2022, 82, 190–208.e17.
- Lustig, L.R.; Kumar, A.S.; Schwämmle, T.; Dunkel, I.; Noviello, G.; Limberg, E.; Weigert, R.; Pacini, G.; Buschow, R.; Ghauri, A.; et al. GATA Transcription Factors Drive Initial Xist Upregulation after Fertilization through Direct Activation of Long-Range Enhancers. Nat. Cell Biol. 2023, 25, 1704–1715.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.0K
Revisions:
2 times
(View History)
Update Date:
23 Feb 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




